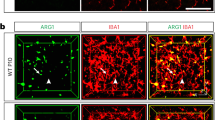
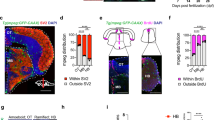
Abstract
The embryonic ectoderm development (EED) is a core component of the polycomb-repressive complex 2 (PRC2) whose mutations are linked to neurodevelopmental abnormalities, intellectual disability, and neurodegeneration. Although EED has been extensively studied in neural stem cells and oligodendrocytes, its role in microglia is incompletely understood. Here, we show that microglial EED is essential for synaptic pruning during the postnatal stage of brain development. The absence of microglial EED at early postnatal stages resulted in reduced spines and impaired synapse density in the hippocampus at adulthood, accompanied by upregulated expression of phagocytosis-related genes in microglia. As a result, deletion of microglial Eed impaired hippocampus-dependent learning and memory in mice. These results suggest that microglial EED is critical for normal synaptic and cognitive functions during postnatal development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The microglia RNA-seq data have been deposited in the Genome Sequence Archive in the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences with accession number CRA006251.
References
Liu PP, Xu YJ, Dai SK, Du HZ, Wang YY, Li XG, et al. Polycomb protein EED regulates neuronal differentiation through targeting SOX11 in hippocampal dentate Gyrus. Stem Cell Rep. 2019;13:115–31.
Ueda T, Nakata Y, Nagamachi A, Yamasaki N, Kanai A, Sera Y, et al. Propagation of trimethylated H3K27 regulated by polycomb protein EED is required for embryogenesis, hematopoietic maintenance, and tumor suppression. Proc Natl Acad Sci USA. 2016;113:10370–5.
Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7.
Ueda T, Sanada M, Matsui H, Yamasaki N, Honda ZI, Shih LY, et al. EED mutants impair polycomb repressive complex 2 in myelodysplastic syndrome and related neoplasms. Leukemia. 2012;26:2557–60.
Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G. Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 1999;13:2691–703.
Richie ER, Schumacher A, Angel JM, Holloway M, Rinchik EM, Magnuson T. The polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene. 2002;21:299–306.
Sauvageau M, Miller M, Lemieux S, Lessard J, Hebert J, Sauvageau G. Quantitative expression profiling guided by common retroviral insertion sites reveals novel and cell type specific cancer genes in leukemia. Blood. 2008;111:790–9.
Cooney E, Bi W, Schlesinger AE, Vinson S, Potocki L. Novel EED mutation in patient with Weaver syndrome. Am J Med Genet A. 2017;173:541–5.
Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, et al. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90:110–8.
Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;384:648.
Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–13.
Wang J, Yang L, Dong C, Wang J, Xu L, Qiu Y, et al. EED-mediated histone methylation is critical for CNS myelination and remyelination by inhibiting WNT, BMP, and senescence pathways. Sci Adv. 2020;6:eaaz6477.
Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. 2018;18:225–42.
Madore C, Yin Z, Leibowitz J, Butovsky O. Microglia, lifestyle stress, and neurodegeneration. Immunity. 2020;52:222–40.
Nayak D, Roth TL, McGavern DB. Microglia development and function. Annu Rev Immunol. 2014;32:367–402.
Masuda T, Sankowski R, Staszewski O, Bottcher C, Amann L, Sagar, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–92.
Doorn KJ, Breve JJ, Drukarch B, Boddeke HW, Huitinga I, Lucassen PJ, et al. Brain region-specific gene expression profiles in freshly isolated rat microglia. Front Cell Neurosci. 2015;9:84.
Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–23 e210.
Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020;30:1271–81.
Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016;19:504–16.
Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–71 e256.
Tan YL, Yuan Y, Tian L. Microglial regional heterogeneity and its role in the brain. Mol Psychiatry. 2020;25:351–67.
De Biase LM, Bonci A. Region-specific phenotypes of microglia: the role of local regulatory cues. Neuroscientist. 2019;25:314–33.
Ayata P, Badimon A, Strasburger HJ, Duff MK, Montgomery SE, Loh YE, et al. Epigenetic regulation of brain region-specific microglia clearance activity. Nat Neurosci. 2018;21:1049–60.
Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670.
Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. 2014;17:400–6.
Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–8.
Filipello F, Morini R, Corradini I, Zerbi V, Canzi A, Michalski B, et al. The microglial innate immune receptor TREM2 is required for synapse elimination and normal brain connectivity. Immunity. 2018;48:979–91 e978.
Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–90 e1217.
Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–81 e569.
Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–40.
Griciuc A, Patel S, Federico AN, Choi SH, Innes BJ, Oram MK, et al. TREM2 acts downstream of CD33 in modulating microglial pathology in Alzheimer’s disease. Neuron. 2019;103:820–35 e827.
Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat Commun. 2019;10:1679.
Wu Y, Dissing-Olesen L, MacVicar BA, Stevens B. Microglia: dynamic mediators of synapse development and plasticity. Trends Immunol. 2015;36:605–13.
Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci. 2009;29:3974–80.
Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–8.
Wang C, Yue H, Hu Z, Shen Y, Ma J, Li J, et al. Microglia mediate forgetting via complement-dependent synaptic elimination. Science. 2020;367:688–94.
Andoh M, Koyama R. Microglia regulate synaptic development and plasticity. Dev Neurobiol. 2021;81:568–90.
McCarthy MM. Location, location, location: microglia are where they live. Neuron. 2017;95:233–5.
Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–905.
Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–40.
Peng H, Geil Nickell CR, Chen KY, McClain JA, Nixon K. Increased expression of M1 and M2 phenotypic markers in isolated microglia after four-day binge alcohol exposure in male rats. Alcohol. 2017;62:29–40.
Kobayashi K, Imagama S, Ohgomori T, Hirano K, Uchimura K, Sakamoto K, et al. Minocycline selectively inhibits M1 polarization of microglia. Cell Death Dis. 2013;4:e525.
Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–5.
Nikodemova M, Kimyon RS, De I, Small AL, Collier LS, Watters JJ. Microglial numbers attain adult levels after undergoing a rapid decrease in cell number in the third postnatal week. J Neuroimmunol. 2015;278:280–8.
Eich ML, Athar M, Ferguson JE 3rd, Varambally S. EZH2-targeted therapies in cancer: hype or a reality. Cancer Res. 2020;80:5449–58.
Laugesen A, Hojfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell. 2019;74:8–18.
Zhang F, Yan Y, Cao X, Zhang J, Li Y, Guo C. Methylation of microRNA-338-5p by EED promotes METTL3-mediated translation of oncogene CDCP1 in gastric cancer. Aging. 2021;13:12224–38.
Klus P, Cirillo D, Botta Orfila T, Gaetano Tartaglia G. Neurodegeneration and cancer: where the disorder prevails. Sci Rep. 2015;5:15390.
Lanni C, Masi M, Racchi M, Govoni S. Cancer and Alzheimer’s disease inverse relationship: an age-associated diverging derailment of shared pathways. Mol Psychiatry. 2021;26:280–95.
Musicco M, Adorni F, Di Santo S, Prinelli F, Pettenati C, Caltagirone C, et al. Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study. Neurology. 2013;81:322–8.
Ibanez K, Boullosa C, Tabares-Seisdedos R, Baudot A, Valencia A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 2014;10:e1004173.
Cheray M, Stratoulias V, Joseph B, Grabert K. The rules of engagement: do microglia seal the fate in the inverse relation of Glioma and Alzheimer’s disease? Front Cell Neurosci. 2019;13:522.
Liu PP, Tang GB, Xu YJ, Zeng YQ, Zhang SF, Du HZ, et al. MiR-203 interplays with polycomb repressive complexes to regulate the proliferation of neural stem/progenitor cells. Stem Cell Rep. 2017;9:190–202.
Dai SK, Liu PP, Du HZ, Liu X, Xu YJ, Liu C, et al. Histone crotonylation regulates neural stem cell fate decisions by activating bivalent promoters. EMBO Rep. 2021; 22:e52023.
Tang GB, Zeng YQ, Liu PP, Mi TW, Zhang SF, Dai SK, et al. The histone H3K27 demethylase UTX regulates synaptic plasticity and cognitive behaviors in mice. Front Mol Neurosci. 2017;10:267.
Cheng Y, Wang ZM, Tan W, Wang X, Li Y, Bai B, et al. Partial loss of psychiatric risk gene Mir137 in mice causes repetitive behavior and impairs sociability and learning via increased Pde10a. Nat Neurosci. 2018;21:1689–703.
Saura J, Tusell JM, Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–9.
Gao X, Enikolopov G, Chen J. Moderate traumatic brain injury promotes proliferation of quiescent neural progenitors in the adult hippocampus. Exp Neurol. 2009;219:516–23.
Acknowledgements
We appreciate the editors and five anonymous reviewers for the in-depth comments, suggestions, and corrections, which have greatly improved the paper. This work was supported by the National Key Research and Development Program of China Project (2018YFA0108001/2021YFA1101402), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010300), the National Science Foundation of China (32170808), and the Open Project Program of State Key Laboratory of Stem Cell and Reproductive Biology.
Author information
Authors and Affiliations
Contributions
ZQT, CML, and YYW, conceived; ZQT, CML, and YT, supervised this study; YYW, ZQT, and CML, designed experiments; YYW, YSD, SKD, TWM, PPL, CL, RYL, BDH, XCH, HZD, and HCY, performed and analyzed experiments; SKD and YYW, performed bioinformatics analysis. YYW and ZQT, prepared figures and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, YY., Deng, YS., Dai, SK. et al. Loss of microglial EED impairs synapse density, learning, and memory. Mol Psychiatry 27, 2999–3009 (2022). https://doi.org/10.1038/s41380-022-01576-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01576-w