Abstract
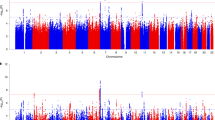
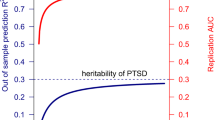
UK Biobank (UKB) is a key contributor in mental health genome-wide association studies (GWAS) but only ~31% of participants completed the Mental Health Questionnaire (“MHQ responders”). We predicted generalized anxiety disorder (GAD), posttraumatic stress disorder (PTSD), and major depression symptoms using elastic net regression in the ~69% of UKB participants lacking MHQ data (“MHQ non-responders”; NTraining = 50%; NTest = 50%), maximizing the informative sample for these traits. MHQ responders were more likely to be female, from higher socioeconomic positions, and less anxious than non-responders. Genetic correlation of GAD and PTSD between MHQ responders and non-responders ranged from 0.636 to 1.08; both were predicted by polygenic scores generated from independent cohorts. In meta-analyses of GAD (N = 489,579) and PTSD (N = 497,803), we discovered many novel genomic risk loci (13 for GAD and 40 for PTSD). Transcriptomic analyses converged on altered regulation of prenatal dorsolateral prefrontal cortex in these disorders. Our results provide one roadmap by which sample size and statistical power may be improved for gene discovery of incompletely ascertained traits in the UKB and other biobanks with limited mental health assessment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data used to generate figures for this study are provided as Supplementary Material. Elastic net weights are provided as Supplementary Material. GWAS summary data are accessible at 10.5281/zenodo.4767570. This research has been conducted using the UK Biobank Resource (application reference no. 58146) and is available to bona fide researchers through approved access. Out-sample polygenic risk scoring utilized the Yale-Penn cohort (dbGaP Study Accession: phs000425.v1.p1) and the Philadelphia Neurodevelopmental Cohort (dbGaP Study Accession: phs000607.v3.p2). The dbGAP data used herein are available for approved access download from dbGAP data request portal.
Code availability
Previously developed pipelines were used to produce the results for this study. No custom code was developed to generate the data used to draw any of our conclusions.
References
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Levey DF, Gelernter J, Polimanti R, Zhou H, Cheng Z, Aslan M, et al. Reproducible genetic risk loci for anxiety: results from approximately 200,000 participants in the Million Veteran Program. Am J Psychiatry. 2020;177:223–32.
Levey DF, Stein MB, Wendt FR, Pathak GA, Zhou H, Aslan M, et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci. 2021;24:954–63.
Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10:4558.
Stein MB, Levey DF, Cheng Z, Wendt FR, Harrington K, Pathak GA, et al. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nat Genet. 2021;53:174–84.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
The All of Us Research Program Investigators. The “All of Us” Research Program. N Engl J Med. 2019;381:668–76.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9.
Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–23.
Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, et al. Overview of the BioBank Japan Project: study design and profile. J Epidemiol. 2017;27:S2–8.
Davis KAS, Coleman JRI, Adams M, Allen N, Breen G, Cullen B, et al. Mental health in UK Biobank—development, implementation and results from an online questionnaire completed by 157 366 participants: a reanalysis. BJPsych Open. 2020;6:e18.
Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry. 2020;25:3292–3303.
Lebovitch DS, Johnson JS, Dueñas HR, Huckins LM. Phenotype risk scores: moving beyond ‘cases’ and ‘controls’ to classify psychiatric disease in hospital-based biobanks. medRxiv. 2021. https://doi.org/10.1101/2021.01.25.21249615.
Kroenke K, Spitzer RL, Williams JB, Löwe B. The Patient Health Questionnaire Somatic, Anxiety, and Depressive Symptom Scales: a systematic review. Gen Hosp Psychiatry. 2010;32:345–59.
Lang AJ, Stein MB. An abbreviated PTSD checklist for use as a screening instrument in primary care. Behav Res Ther. 2005;43:585–94.
Levey DF, Gelernter J, Polimanti R, Zhou H, Cheng Z, Aslan M, et al. Reproducible genetic risk loci for anxiety: results from approximately 200,000 participants in the Million Veteran Program. Am J Psychiatry. 2020. https://doi.org/10.1176/appi.ajp.2019.19030256.
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1.
Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–37.
Kessler RC, Aguilar-Gaxiola S, Alonso J, Chatterji S, Lee S, Ormel J, et al. The global burden of mental disorders: an update from the WHO World Mental Health (WMH) surveys. Epidemiol Psichiatr Soc. 2009;18:23–33.
Yehuda R, Hoge CW, McFarlane AC, Vermetten E, Lanius RA, Nievergelt CM, et al. Post-traumatic stress disorder. Nat Rev Dis Prim. 2015;1:15057.
Hasin DS, Sarvet AL, Meyers JL, Saha TD, Ruan WJ, Stohl M, et al. Epidemiology of adult DSM-5 major depressive disorder and its specifiers in the United States. JAMA Psychiatry. 2018;75:336–46.
Choi SW, O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8:giz082.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Watanabe K, Umicevic Mirkov M, de Leeuw CA, van den Heuvel MP, Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat Commun. 2019;10:3222.
The GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30.
Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:eaat7615.
Sey NYA, Hu B, Mah W, Fauni H, McAfee JC, Rajarajan P, et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat Neurosci. 2020;23:583–93.
Wang G, Sarkar A, Carbonetto P, Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J R Stat Soc: Ser B. 2020;82:1273–1300.
Wu Y, Byrne EM, Zheng Z, Kemper KE, Yengo L, Mallett AJ, et al. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun. 2019;10:1891.
O’Connor LJ, Price AL. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet. 2018;50:1728–34.
Napolitano F, Carrella D, Mandriani B, Pisonero-Vaquero S, Sirci F, Medina DL, et al. gene2drug: a computational tool for pathway-based rational drug repositioning. Bioinformatics. 2018;34:1498–505.
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35.
Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–9.
Uçar MK, Nour M, Sindi H, Polat K. The effect of training and testing process on machine learning in biomedical datasets. Math Probl Eng. 2020;2020:2836236.
Wendt FR, Pathak GA, Lencz T, Krystal JH, Gelernter J, Polimanti R. Multivariate genome-wide analysis of education, socioeconomic status, and brain phenome. Nat Hum Behav. 2020. https://doi.org/10.1038/s41562-41020-00980-y.
Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224.
Walsh RM, Shen EY, Bagot RC, Anselmo A, Jiang Y, Javidfar B, et al. Phf8 loss confers resistance to depression-like and anxiety-like behaviors in mice. Nat Commun. 2017;8:15142.
Kim HJ, Hur SW, Park JB, Seo J, Shin JJ, Kim SY, et al. Histone demethylase PHF2 activates CREB and promotes memory consolidation. EMBO Rep. 2019;20:e45907.
Lv C, Zhang H, Li Z, Zhang H, Zhang Y, Li R, et al. Low-dose IL-2 therapy compensates for metabolic shifts and reverses anxiety-like behavior in PD-1 deficiency-induced autoimmunity. Cell Mol Immunol. 2021;18:1336–8.
Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, et al. Largest GWAS of PTSD (N 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 2018;23:666–73.
Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG, et al. Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry. 2016;21:1391–9.
Calkins ME, Merikangas KR, Moore TM, Burstein M, Behr MA, Satterthwaite TD, et al. The Philadelphia neurodevelopmental cohort: constructing a deep phenotyping collaborative. J Child Psychol Psychiatry. 2015;56:1356–69.
Robinson EB, Kirby A, Ruparel K, Yang J, McGrath L, Anttila V, et al. The genetic architecture of pediatric cognitive abilities in the Philadelphia neurodevelopmental cohort. Mol Psychiatry. 2015;20:454–8.
Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, et al. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA). Drug Alcohol Depend. 2007;91:85–90.
Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the semi-structured assessment for drug dependence and alcoholism (SSADDA). Drug Alcohol Depend. 2005;80:303–12.
Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–8.
Chen J-J, Bai S-J, Li W-W, Zhou C-J, Zheng P, Fang L, et al. Urinary biomarker panel for diagnosing patients with depression and anxiety disorders. Transl Psychiatry. 2018;8:192.
Meier SM, Trontti K, Purves KL, Als TD, Grove J, Laine M, et al. Genetic variants associated with anxiety and stress-related disorders: a genome-wide association study and mouse-model study. JAMA Psychiatry. 2019;76:924–32.
Gelernter J, Sun N, Polimanti R, Pietrzak R, Levey DF, Bryois J, et al. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat Neurosci. 2019;22:1394–401.
Hill WD, Weiss A, Liewald DC, Davies G, Porteous DJ, Hayward C, et al. Genetic contributions to two special factors of neuroticism are associated with affluence, higher intelligence, better health, and longer life. Mol Psychiatry. 2020;25:3034–52.
Russo AC, Fingerhut EC. Consistency of self-reported neurocognitive symptoms, post-traumatic stress disorder symptoms, and concussive events from end of first deployment to veteran health administration comprehensive traumatic brain injury evaluation by operations enduring freedom/Iraqi Freedom/New Dawn Veterans. Arch Clin Neuropsychol. 2017;32:184–97.
Beesdo K, Knappe S, Pine DS. Anxiety and anxiety disorders in children and adolescents: developmental issues and implications for DSM-V. Psychiatr Clin North Am. 2009;32:483–524.
Aupperle RL, Allard CB, Grimes EM, Simmons AN, Flagan T, Behrooznia M, et al. Dorsolateral prefrontal cortex activation during emotional anticipation and neuropsychological performance in posttraumatic stress disorder. Arch Gen Psychiatry. 2012;69:360–71.
Philip NS, Barredo J, van ‘t Wout-Frank M, Tyrka AR, Price LH, Carpenter LL. Network mechanisms of clinical response to transcranial magnetic stimulation in posttraumatic stress disorder and major depressive disorder. Biol Psychiatry. 2018;83:263–72.
Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9.
Del Giacco SR, Cappai A, Gambula L, Cabras S, Perra S, Manconi PE, et al. The asthma-anxiety connection. Respir Med. 2016;120:44–53.
Mitchell BL, Thorp JG, Wu Y, Campos AI, Nyholt DR, Gordon SD, et al. Polygenic risk scores derived from varying definitions of depression and risk of depression. JAMA Psychiatry. 2021;78:1152–60.
Takeshita J, Wang S, Loren AW, Mitra N, Shults J, Shin DB, et al. Association of racial/ethnic and gender concordance between patients and physicians with patient experience ratings. JAMA Netw Open. 2020;3:e2024583.
Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–6.
Acknowledgements
This research has been conducted using the UK Biobank Resource (application reference no. 58146). The authors thank the research participants and employees of the UK Biobank for making this work possible. This study was supported by National Institutes of Health (R21 DC018098, R21 DA047527, R33 DA047527, and F32 MH122058) and a Faculty Scholar Award from the Seaver Foundation: “Analytical Genomics of Vulnerable Populations.” The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
FRW and RP conceived the study design; FRW performed phenotype prediction, demographic comparisons, multi-trait conditioning, genome-wide association study meta-analyses, causal inference analysis, functional annotation, and PRS in the PNC; GAP performed fine mapping; JDD assisted with multi-trait conditioning; FDA performed drug repurposing analyses; DK performed genetic correlation; BCM performed GWAS statistics-level PRS; FRW, GAP, JDD, DSL, DFL, MBS, HRK, KCK, JG, LMH, and RP contributed to data interpretation; FRW, GAP and RP contributed to data visualization and presentation; FRW drafted the original manuscript. All authors critically evaluated and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
HRK is a member of an advisory board for Dicerna Pharmaceuticals, a consultant to Sophrosyne Pharmaceuticals, a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which for the past 3 years was supported by AbbVie, Alkermes, Amygdala Neurosciences, Arbor, DIicerna, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer, and is paid for his editorial work on the journal Alcoholism: Clinical and Experimental Research. HRK and JG are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. MBS is paid for his editorial work on the journals Biological Psychiatry and Depression and Anxiety, and the health professional reference Up-To-Date; he has also in the past 3 years received consulting income from Actelion, Acadia Pharmaceuticals, Aptinyx, Bionomics, BioXcel Therapeutics, Clexio, EmpowerPharm, GW Pharmaceuticals, Janssen, Jazz Pharmaceuticals, and Roche/Genentech, and has stock options in Oxeia Biopharmaceuticals and Epivario. RP and JG are paid for their editorial work on the journal Complex Psychiatry. The other authors have no competing interests to report.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wendt, F.R., Pathak, G.A., Deak, J.D. et al. Using phenotype risk scores to enhance gene discovery for generalized anxiety disorder and posttraumatic stress disorder. Mol Psychiatry 27, 2206–2215 (2022). https://doi.org/10.1038/s41380-022-01469-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01469-y
This article is cited by
-
Effects of sex and gender on the etiologies and presentation of select internalizing psychopathologies
Translational Psychiatry (2024)
-
The impact of assortative mating, participation bias and socioeconomic status on the polygenic risk of behavioural and psychiatric traits
Nature Human Behaviour (2024)
-
Genome-wide association study in 404,302 individuals identifies 7 significant loci for reaction time variability
Molecular Psychiatry (2023)
-
Functional and molecular characterization of suicidality factors using phenotypic and genome-wide data
Molecular Psychiatry (2023)
-
Participation bias in the UK Biobank distorts genetic associations and downstream analyses
Nature Human Behaviour (2023)