Abstract
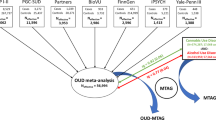
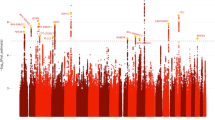
Substance use disorders (SUDs) are moderately to highly heritable and are in part cross-transmitted genetically, as observed in twin and family studies. We performed exome-focused genotyping to examine the cross-transmission of four SUDs: alcohol use disorder (AUD, n = 4487); nicotine use disorder (NUD, n = 4394); cannabis use disorder (CUD, n = 954); and nonmedical prescription opioid use disorder (NMPOUD, n = 346) within a large nationally representative sample (n = 36,309), the National Epidemiologic Survey on Alcohol and Related Conditions-III (NESARC-III). All diagnoses were based on in-person structured psychiatric interview (AUDADIS-5). SUD cases were compared alone and together to 3959 “super controls” who had neither a SUD nor a psychiatric disorder using an exome-focused array assaying 363,496 SNPs, yielding a representative view of within-disorder and cross-disorder genetic influences on SUDs. The 29 top susceptibility genes for one or more SUDs overlapped highly with genes previously implicated by GWAS of SUD. Polygenic scores (PGS) were computed within the European ancestry (EA) component of the sample (n = 12,505) using summary statistics from each of four clinically distinct SUDs compared to the 3959 “super controls” but then used for two distinctly different purposes: to predict SUD severity (mild, moderate, or severe) and to predict each of the other 3 SUDs. Our findings based on PGS highlight shared and unshared genetic contributions to the pathogenesis of SUDs, confirming the strong cross-inheritance of AUD and NUD as well as the distinctiveness of inheritance of opioid use disorder.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The individual-level genetic data (22,848 samples) with phenotypic variables (n = 4,320) for NESARC-III (family history, adverse childhood experiences, substance use, mood, anxiety, personality and posttraumatic stress disorders) are available in dbGaP (accession: phs001590.v2.p1). [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001590.v2.p1].
References
Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382:1575–86.
Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003;60:929–37.
Ducci F, Goldman D. The genetic basis of addictive disorders. Psychiatr Clin North Am. 2012;35:495–519.
Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–32.
Goldman D, Bergen A. General and specific inheritance of substance abuse and alcoholism. Arch Gen Psychiatry. 1998;55:964–5.
Cheng Z, Zhou H, Sherva R, Farrer LA, Kranzler HR, Gelernter J. Genome-wide association study identifies a regulatory variant of RGMA associated with opioid dependence in European Americans. Biol Psychiatry. 2018;84:762–70.
Li D, Zhao H, Kranzler HR, Li MD, Jensen KP, Zayats T, et al. Genome-wide association study of copy number variations (CNVs) with opioid dependence. Neuropsychopharmacology. 2015;40:1016–26.
Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44.
Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Adams MJ, Howard DM, et al. Genome-wide association study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2019;176:107–18.
Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, et al. Genome-wide association study of cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry. 2016;73:472–80.
Stringer S, Minică CC, Verweij KJ, Mbarek H, Bernard M, Derringer J, et al. Genome-wide association study of lifetime cannabis use based on a large meta-analytic sample of 32,330 subjects from the International Cannabis Consortium. Transl Psychiatry. 2016;6:e769.
Tobacco, Genetics C. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–7.
Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21:1656–69.
Goldman D. Polygenic risk scores in psychiatry. Biol Psychiatry. 2017;82:698–9.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Grant BF, Goldstein RB, Smith SM, Jung J, Zhang H, Chou SP, et al. The Alcohol Use Disorder and Associated Disabilities Interview Schedule-5 (AUDADIS-5): reliability of substance use and psychiatric disorder modules in a general population sample. Drug Alcohol Depend. 2015;148:27–33.
Grant BF, Chu, A, Sigman, R, Amsbary, M, Kali, J, Sugawara Y, et al. Source and Accuracy Statement: National Epidemiologic Survey on Alcohol and Related Conditions-III (NESARC-III) [https://www.niaaa.nih.gov/sites/default/files/NESARC_Final_Report_FINAL_1_8_15.pdf]. National Institute on Alcohol Abuse and Alcoholism. 2014.
Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, et al. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry. 2017;22:1359–67.
Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10:1499.
Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry. 2017;22:1376–84.
Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42.
Chou SP, Goldstein RB, Smith SM, Huang B, Ruan WJ, Zhang H, et al. The epidemiology of DSM-5 nicotine use disorder: results from the national epidemiologic survey on alcohol and related conditions-III. J Clin Psychiatry. 2016;77:1404–12.
Grant BF, Chou SP, Saha TD, Pickering RP, Kerridge BT, Ruan WJ, et al. Prevalence of 12-month alcohol use, high-risk drinking, and DSM-IV alcohol use disorder in the United States, 2001-2 to 2012-3: Results From the National Epidemiologic Survey on Alcohol and Related Conditions. JAMA Psychiatry. 2017;74:911–23.
Grant BF, Saha TD, Ruan WJ, Goldstein RB, Chou SP, Jung J, et al. Epidemiology of DSM-5 drug use disorder: results from the national epidemiologic survey on alcohol and related conditions-III. JAMA Psychiatry. 2016;73:39–47.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–6.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
Feng S, Liu D, Zhan X, Wing MK, Abecasis GR. RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics. 2014;30:2828–9.
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7.
Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–37.
van der Sluis S, Posthuma D, Dolan CV. TATES: efficient multivariate genotype-phenotype analysis for genome-wide association studies. PLoS Genet. 2013;9:e1003235.
Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics 2015;31:1466–8.
Song W, Kossowsky J, Torous J, Chen CY, Huang H, Mukamal KJ, et al. Genome-wide association analysis of opioid use disorder: A novel approach using clinical data. Drug Alcohol Depend. 2020;217:108276.
Watanabe K, Stringer S, Frei O, Umicevic Mirkov M, de Leeuw C, Polderman TJC, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51:1339–48.
Yu C, McClellan J. Genetics of substance use disorders. Child Adolesc Psychiatr Clin N. Am. 2016;25:377–85.
Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–15.
Zhou H, Rentsch CT, Cheng Z, Kember RL, Nunez YZ, Sherva RM, et al. Association of OPRM1 functional coding variant with opioid use disorder: a genome-wide association study. JAMA Psychiatry. 2020;77:1072–80.
Demontis D, Rajagopal VM, Thorgeirsson TE, Als TD, Grove J, Leppala K, et al. Genome-wide association study implicates CHRNA2 in cannabis use disorder. Nat Neurosci. 2019;22:1066–74.
Panjwani N, Wang F, Mastromatteo S, Bao A, Wang C, He G, et al. LocusFocus: Web-based colocalization for the annotation and functional follow-up of GWAS. PLoS Comput Biol. 2020;16:e1008336.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Zhou H, Sealock JM, Sanchez-Roige S, Clarke TK, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23:809–18.
Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10:3328.
Heath AC, Martin NG. Genetic models for the natural history of smoking: evidence for a genetic influence on smoking persistence. Addict Behav. 1993;18:19–34.
Han C, McGue MK, Iacono WG. Lifetime tobacco, alcohol and other substance use in adolescent Minnesota twins: univariate and multivariate behavioral genetic analyses. Addiction. 1999;94:981–93.
Hicks BM, Krueger RF, Iacono WG, McGue M, Patrick CJ. Family transmission and heritability of externalizing disorders: a twin-family study. Arch Gen Psychiatry. 2004;61:922–8.
Kandel DB, Kandel ER. A molecular basis for nicotine as a gateway drug. N Engl J Med. 2014;371:2038–9.
Kwako LE, Momenan R, Litten RZ, Koob GF, Goldman D. Addictions neuroclinical assessment: a neuroscience-based framework for addictive disorders. Biol Psychiatry. 2016;80:179–89.
Kwako LE, Schwandt ML, Ramchandani VA, Diazgranados N, Koob GF, Volkow ND, et al. Neurofunctional domains derived from deep behavioral phenotyping in alcohol use disorder. Am J Psychiatry. 2019;176:744–53.
Acknowledgements
We thank the study participants and their families. The National Epidemiologic Survey on Alcohol and Related Conditions-III (NESARC-III) is funded by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) with supplemental support from the National Institute on Drug Abuse. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Author information
Authors and Affiliations
Contributions
Study conception and design: HZ, DG, SPC; Acquisition of data: BFG, HZ, SPC, WJR, BTK, BH, TDS, AZF, VW, JJ; Analysis and interpretation of data: HZ, DG, SPC, CAH; Drafting manuscript: HZ, DG, SPC; Administrative & technical functions: BFG, HZ, SPC; Clerical & material support: VW; Comments & discussion: AP, CAH.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, H., Grant, B.F., Hodgkinson, C.A. et al. Strong and weak cross-inheritance of substance use disorders in a nationally representative sample. Mol Psychiatry 27, 1742–1753 (2022). https://doi.org/10.1038/s41380-021-01370-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01370-0