Abstract
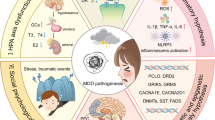
Major depressive disorder (MDD) is a brain disorder often characterized by recurrent episode and remission phases. The molecular correlates of MDD have been investigated in case-control comparisons, but the biological alterations associated with illness trait (regardless of clinical phase) or current state (symptomatic and remitted phases) remain largely unknown, limiting targeted drug discovery. To characterize MDD trait- and state-dependent changes, in single or recurrent depressive episode or remission, we generated transcriptomic profiles of subgenual anterior cingulate cortex of postmortem subjects in first MDD episode (n = 20), in remission after a single episode (n = 15), in recurrent episode (n = 20), in remission after recurring episodes (n = 15) and control subject (n = 20). We analyzed the data at the gene, biological pathway, and cell-specific molecular levels, investigated putative causal events and therapeutic leads. MDD-trait was associated with genes involved in inflammation, immune activation, and reduced bioenergetics (q < 0.05) whereas MDD-states were associated with altered neuronal structure and reduced neurotransmission (q < 0.05). Cell-level deconvolution of transcriptomic data showed significant change in density of GABAergic interneurons positive for corticotropin-releasing hormone, somatostatin, or vasoactive-intestinal peptide (p < 3 × 10−3). A probabilistic Bayesian-network approach showed causal roles of immune-system-activation (q < 8.67 × 10−3), cytokine-response (q < 4.79 × 10−27) and oxidative-stress (q < 2.05 × 10−3) across MDD-phases. Gene-sets associated with these putative causal changes show inverse associations with the transcriptomic effects of dopaminergic and monoaminergic ligands. The study provides first insights into distinct cellular and molecular pathologies associated with trait- and state-MDD, on plasticity mechanisms linking the two pathologies, and on a method of drug discovery focused on putative disease-causing pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All datasets analyzed during the current study are available as supplementary tables. Raw data (count matrix, fastq.gz, or.bam) are available from the corresponding author on reasonable request.
References
Malhi GS, Mann JJ. Depression. Lancet (Lond, Engl). 2018;392:2299–312.
Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, et al. Major depressive disorder. Nat Rev Dis Prim. 2016;2:16065.
Sibille E, French B. Biological substrates underpinning diagnosis of major depression. Int J Neuropsychopharmacol. 2013. 2013. https://doi.org/10.1017/S1461145713000436.
Scharnowski F, Nicholson AA, Pichon S, Rosa MJ, Rey G, Eickhoff SB, et al. The role of the subgenual anterior cingulate cortex in dorsomedial prefrontal–amygdala neural circuitry during positive-social emotion regulation. Hum Brain Mapp. 2020. 2020. https://doi.org/10.1002/hbm.25001.
Haas BW, Omura K, Constable RT, Canli T. Emotional conflict and neuroticism: personality-dependent activation in the amygdala and subgenual anterior cingulate. Behav Neurosci. 2007. 2007. https://doi.org/10.1037/0735-7044.121.2.249.
Masten CL, Eisenberger NI, Borofsky LA, Mcnealy K, Pfeifer JH, Dapretto M. Subgenual anterior cingulate responses to peer rejection: a marker of adolescents’ risk for depression. Dev Psychopathol. 2011. 2011. https://doi.org/10.1017/S0954579410000799.
Mayberg HS, Liotti M, Brannan SK, McGinnis S, Mahurin RK, Jerabek PA, et al. Reciprocal limbic-cortical function and negative mood: converging PET findings in depression and normal sadness. Am J Psychiatry. 1999. 1999.
Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, et al. Deep brain stimulation for treatment-resistant depression. Neuron 2005;45:651–60.
Merkl A, Neumann WJ, Huebl J, Aust S, Horn A, Krauss JK, et al. Modulation of beta-band activity in the subgenual anterior cingulate cortex during emotional empathy in treatment-resistant depression. Cereb Cortex. 2016. 2016. https://doi.org/10.1093/cercor/bhv100.
Hasler G, van der Veen JW, Tumonis T, Meyers N, Shen J, Drevets WC. Reduced Prefrontal Glutamate/Glutamine and γ-Aminobutyric Acid Levels in Major Depression Determined Using Proton Magnetic Resonance Spectroscopy. Arch Gen Psychiatry. 2007;64:193.
Sanacora G, Mason GF, Rothman DL, Behar KL, Hyder F, Petroff OAC, et al. Reduced cortical γ-aminobutyric acid levels in depressed patients determined by proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 1999. 1999. https://doi.org/10.1001/archpsyc.56.11.1043.
Sanacora G, Mason GF, Rothman DL, Krystal JH. Increased occipital cortex GABA concentrations in depressed patients after therapy with selective serotonin reuptake inhibitors. Am J Psychiatry. 2002. 2002. https://doi.org/10.1176/appi.ajp.159.4.663.
Sanacora G, Rothman DL, Mason G, Krystal JH. Clinical Studies Implementing Glutamate Neurotransmission in Mood Disorders. Ann N Y Acad Sci. 2003;1003:292–308.
Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, et al. Global brain gene expression analysis links Glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009. 2009. https://doi.org/10.1371/journal.pone.0006585.
Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, et al. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int J Neuropsychopharmacol. 2013. 2013. https://doi.org/10.1017/S1461145712000016.
Ongur D, Drevets WC, Price JL. Glial reduction in the subgenual prefrontal cortex in mood disorders. Proc Natl Acad Sci. 2002. 2002. https://doi.org/10.1073/pnas.95.22.13290.
Medina A, Watson SJ, Bunney W, Myers RM, Schatzberg A, Barchas J, et al. Evidence for alterations of the glial syncytial function in major depressive disorder. J Psychiatr Res. 2016. 2016. https://doi.org/10.1016/j.jpsychires.2015.10.010.
Sibille E, Morris HM, Kota RS, Lewis DA. GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. Int J Neuropsychopharmacol. 2011;14:721–34.
Tripp A, Kota RS, Lewis DA, Sibille E. Reduced somatostatin in subgenual anterior cingulate cortex in major depression. Neurobiol Dis. 2011;42:116–24.
Tripp A, Oh H, Guilloux J-PP, Martinowich K, Lewis DA, Sibille E. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. Am J Psychiatry. 2012;169:1194–202.
Sibille E, Wang Y, Joeyen-Waldorf J, Gaiteri C, Surget A, Oh S, et al. A molecular signature of depression in the amygdala. Am J Psychiatry. 2009;166:1011–24.
Scifo E, Pabba M, Kapadia F, Ma T, Lewis DA, Tseng GC, et al. Sustained molecular pathology across episodes and remission in major depressive disorder. Biol Psychiatry. 2018. 2018. https://doi.org/10.1016/j.biopsych.2017.08.008.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Pantazatos SP, Huang Y-Y, Rosoklija GB, Dwork AJ, Arango V, Mann JJ. Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: evidence for altered glial, endothelial and ATPase activity. Mol Psychiatry. 2017;22:760–73.
Ramaker RC, Bowling KM, Lasseigne BN, Hagenauer MH, Hardigan AA, Davis NS, et al. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017. 2017. https://doi.org/10.1186/s13073-017-0458-5.
Labonté B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. Sex-specific transcriptional signatures in human depression. Nat Med. 2017. 2017. https://doi.org/10.1038/nm.4386.
Ding Y, Chang L-C, Wang X, Guilloux J-P, Parrish J, Oh H, et al. Molecular and genetic characterization of depression: overlap with other psychiatric disorders and aging. Mol Neuropsychiatry. 2015;1:1–12.
Bioconductor - GeneOverlap. https://bioconductor.org/packages/release/bioc/html/GeneOverlap.html. Accessed 22 April 2021.
Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016. 2016. https://doi.org/10.1016/j.cels.2016.08.011.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019. 2019. https://doi.org/10.1016/j.cell.2019.05.031.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. Cell type–specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–9.
Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: State of the art. J Biopharm Stat. 2010.
Agrahari R, Foroushani A, Docking TR, Chang L, Duns G, Hudoba M, et al. Applications of Bayesian network models in predicting types of hematological malignancies. Sci Rep. 2018. 2018. https://doi.org/10.1038/s41598-018-24758-5.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15.
Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4.
Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54.
Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013. 2013. https://doi.org/10.1016/j.cell.2013.03.030.
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017. 2017. https://doi.org/10.1016/j.cell.2017.10.049.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019. 20 May 2019. https://doi.org/10.1093/nar/gkz382.
Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010. 2010. https://doi.org/10.1038/nmeth.1439.
Foroushani A, Agrahari R, Docking R, Chang L, Duns G, Hudoba M, et al. Large-scale gene network analysis reveals the significance of extracellular matrix pathway and homeobox genes in acute myeloid leukemia: An introduction to the Pigengene package and its applications. BMC Med Genomics. 2017. 2017. https://doi.org/10.1186/s12920-017-0253-6.
Anisman H, Merali Z. Cytokines, stress and depressive illness: Brain-immune interactions. Ann Med. 2003;35:2–11.
Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112:7285–90.
He Z, Han D, Efimova O, Guijarro P, Yu Q, Oleksiak A, et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat Neurosci. 2017. 2017. https://doi.org/10.1038/nn.4548.
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929–35.
Gross J, Turecki G. Suicide and the Polyamine System. CNS Neurol Disord - Drug Targets. 2013. 2013. https://doi.org/10.2174/18715273113129990095.
Miller AH, Raison CL. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol. 2016;16:22–34.
Wang JQ, Mao L. The ERK pathway: molecular mechanisms and treatment of depression. Mol Neurobiol. 2019;56:6197–205.
Liu CH, Zhang GZ, Li B, Li M, Woelfer M, Walter M, et al. Role of inflammation in depression relapse. J Neuroinflammation. 2019;16:90.
E. Leonard B. The Concept of Depression as a Dysfunction of the Immune System. Curr Immunol Rev. 2010. 2010. https://doi.org/10.2174/157339510791823835.
Warner-Schmidt JL, Duman RS. VEGF as a potential target for therapeutic intervention in depression. Curr Opin Pharmacol. 2008;8:14–9.
Allen J, Romay-Tallon R, Brymer KJ, Caruncho HJ, Kalynchuk LE. Mitochondria and mood: Mitochondrial dysfunction as a key player in the manifestation of depression. Front Neurosci. 2018;12:386.
Culmsee C, Michels S, Scheu S, Arolt V, Dannlowski U, Alferink J. Mitochondria, microglia, and the immune system — How are they linked in affective disorders? Front Psychiatry. 2019. 2019. https://doi.org/10.3389/fpsyt.2018.00739.
Czarny P, Wigner P, Galecki P, Sliwinski T. The interplay between inflammation, oxidative stress, DNA damage, DNA repair and mitochondrial dysfunction in depression. Prog Neuro-PsychopharmacologyBiol Psychiatry. 2018;80:309–21.
Duman RS, Sanacora G, Krystal JH. Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron. 2019;102:75–90.
Banasr M, Dwyer JM, Duman RS. Cell atrophy and loss in depression: reversal by antidepressant treatment. Curr Opin Cell Biol. 2011;23:730–7.
Rajkowska G, Miguel-Hidalgo JJ, Wei J, Dilley G, Pittman SD, Meltzer HY, et al. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry. 1999. 1999. https://doi.org/10.1016/S0006-3223(99)00041-4.
Fee C, Banasr M, Sibille E. Somatostatin-positive gamma-aminobutyric acid interneuron deficits in depression: cortical microcircuit and therapeutic perspectives. Biol Psychiatry. 2017;82:549–59.
Simon RA, Barazanji N, Jones MP, Bednarska O, Icenhour A, Engström M, et al. Vasoactive intestinal polypeptide plasma levels associated with affective symptoms and brain structure and function in healthy females. Sci Rep. 2021;11:1406.
Gjerris A, Rafaelsen OJ, Vendsborg P, Fahrenkrug J, Rehfeld JF. Vasoactive intestinal polypeptide decreased in cerebrospinal fluid (CSF) in atypical depression. Vasoactive intestinal polypeptide, cholecystokinin and gastrin in CSF in psychiatric disorders. J Affect Disord. 1984;7:325–37.
Soria V, Martínez-Amorós È, Escaramís G, Valero J, Pérez-Egea R, García C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and clock and VIP with bipolar disorder. Neuropsychopharmacology 2010;35:1279–89.
Schulz JM, Knoflach F, Hernandez MC, Bischofberger J. Dendrite-targeting interneurons control synaptic NMDA-receptor activation via nonlinear α5-GABAA receptors. Nat Commun. 2018;9:3576.
Prévot T, Sibille E. Altered GABA-mediated information processing and cognitive dysfunctions in depression and other brain disorders. Mol Psychiatry. 2021;26:151–67.
Zaitsev K, Bambouskova M, Swain A, Artyomov MN. Complete deconvolution of cellular mixtures based on linearity of transcriptional signatures. Nat Commun. 2019. 2019. https://doi.org/10.1038/s41467-019-09990-5.
Wang J, Huang M, Torre E, Dueck H, Shaffer S, Murray J, et al. Gene expression distribution deconvolution in single-cell RNA sequencing. Proc Natl Acad Sci USA. 2018. 2018. https://doi.org/10.1073/pnas.1721085115.
Seney ML, Tripp A, McCune S, A. Lewis D, Sibille E. Laminar and cellular analyses of reduced somatostatin gene expression in the subgenual anterior cingulate cortex in major depression. Neurobiol Dis. 2015. 2015. https://doi.org/10.1016/j.nbd.2014.10.005.
Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat Neurosci. 2018. 2018. https://doi.org/10.1038/s41593-018-0154-9.
Meinshausen N, Hauser A, Mooij JM, Peters J, Versteeg P, Bühlmann P. Methods for causal inference from gene perturbation experiments and validation. Proc Natl Acad Sci USA. 2016;113:7361–8.
Acknowledgements
The study was supported by a project grant from the Canadian Institute of Health Research (CIHR) PJT-153175.
Author information
Authors and Affiliations
Contributions
RS and ES conceptualized the study and together wrote the manuscript. RS performed all the sequencing library preparation, quality check and bioinformatics analysis. DFN participated in in-silico validations. TT and AS performed the QPCR validation. HZ participated in Bayesian network analysis. RM participated in cmap analysis and DAL provided the resources.
Corresponding authors
Ethics declarations
Competing interests
ES is founder and Acting Chief Scientific Officer of Damona Pharmaceuticals, a drug development company with small molecules in the pipeline for treatment of cognitive deficits across brain disorders and aging. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shukla, R., Newton, D.F., Sumitomo, A. et al. Molecular characterization of depression trait and state. Mol Psychiatry 27, 1083–1094 (2022). https://doi.org/10.1038/s41380-021-01347-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01347-z
This article is cited by
-
Transcriptional dissection of symptomatic profiles across the brain of men and women with depression
Nature Communications (2023)
-
Differential vulnerability of anterior cingulate cortex cell types to diseases and drugs
Molecular Psychiatry (2022)
-
Antipsychotics impair regulation of glucose metabolism by central glucose
Molecular Psychiatry (2022)
-
Single-cell transcriptome study in forensic medicine: prospective applications
International Journal of Legal Medicine (2022)
-
Strategies to identify candidate repurposable drugs: COVID-19 treatment as a case example
Translational Psychiatry (2021)