Abstract
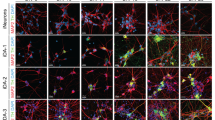
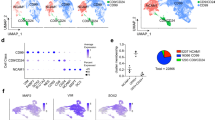
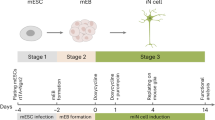
Dopaminergic neurons are critical to movement, mood, addiction, and stress. Current techniques for generating dopaminergic neurons from human induced pluripotent stem cells (hiPSCs) yield heterogenous cell populations with variable purity and inconsistent reproducibility between donors, hiPSC clones, and experiments. Here, we report the rapid (5 weeks) and efficient (~90%) induction of induced dopaminergic neurons (iDANs) through transient overexpression of lineage-promoting transcription factors combined with stringent selection across five donors. We observe maturation-dependent increase in dopamine synthesis and electrophysiological properties consistent with midbrain dopaminergic neuron identity, such as slow-rising after- hyperpolarization potentials, an action potential duration of ~3 ms, tonic sub-threshold oscillatory activity, and spontaneous burst firing at a frequency of ~1.0–1.75 Hz. Transcriptome analysis reveals robust expression of genes involved in fetal midbrain dopaminergic neuron identity. Specifically expressed genes in iDANs, as well as those from isogenic induced GABAergic and glutamatergic neurons, were enriched in loci conferring heritability for cannabis use disorder, schizophrenia, and bipolar disorder; however, each neuronal subtype demonstrated subtype-specific heritability enrichments in biologically relevant pathways, and iDANs alone were uniquely enriched in autism spectrum disorder risk loci. Therefore, iDANs provide a critical tool for modeling midbrain dopaminergic neuron development and dysfunction in psychiatric disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The source data described in this manuscript are available via the PsychENCODE Knowledge Portal (https://psychencode.synapse.org/). The PsychENCODE Knowledge Portal is a platform for accessing data, analyses, and tools generated through grants funded by the National Institute of Mental Health (NIMH) PsychENCODE program. Data is available for general research use according to the following requirements for data access and data attribution: (https://psychencode.synapse.org/DataAccess). For access to content described in this manuscript see: https://doi.org/10.7303/syn25500352.
Code availability
Available from the authors upon request.
References
Schultz W. Multiple dopamine functions at different time courses. Ann Rev Neurosci. 2007. https://doi.org/10.1146/annurev.neuro.28.061604.135722.
Meder D, Herz DM, Rowe JB, Lehéricy S, Siebner HR. The role of dopamine in the brain - lessons learned from Parkinson’s disease. NeuroImage. 2019. https://doi.org/10.1016/j.neuroimage.2018.11.021.
Volkow ND, Wise RA, Baler R. The dopamine motive system: Implications for drug and food addiction. Nat Rev Neurosci. 2017. https://doi.org/10.1038/nrn.2017.130.
Grace AA, Gomes FV. The circuitry of dopamine system regulation and its disruption in schizophrenia: insights into treatment and prevention. Schizophrenia Bull. 2019;45:148–57. https://doi.org/10.1093/schbul/sbx199.
LaMarca EA, Powell SK, Akbarian S, Brennand KJ. Modeling neuropsychiatric and neurodegenerative diseases with induced pluripotent stem cells. Front Pediatrics. 2018;6. https://doi.org/10.3389/fped.2018.00082.
Powell SK, O’Shea CP, Shannon SR, Akbarian S, Brennand KJ. Investigation of schizophrenia with human induced pluripotent stem cells. Adv Neurobiol . 2018;155–206. https://doi.org/10.1007/978-3-030-45493-7_6.
Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011. https://doi.org/10.1038/nature10648.
Addis RC, Hsu FC, Wright RL, Dichter MA, Coulter DA, Gearhart JD. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One. 2011. https://doi.org/10.1371/journal.pone.0028719.
Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011. https://doi.org/10.1038/nature10284.
Mahajani S, Raina A, Fokken C, Kügler S, Bähr M. Homogenous generation of dopaminergic neurons from multiple hiPSC lines by transient expression of transcription factors. Cell Death Dis. 2019. https://doi.org/10.1038/s41419-019-2133-9.
Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011. https://doi.org/10.1073/pnas.1105135108.
Theka I, Caiazzo M, Dvoretskova E, Leo D, Ungaro F, Curreli S, et al. Rapid generation of functional dopaminergic neurons from human induced pluripotent stem cells through a single-step procedure using cell lineage transcription factors. STEM CELLS Transl Med. 2013. https://doi.org/10.5966/sctm.2012-0133.
Beevers JE, Lai MC, Collins E, Booth HDE, Zambon F, Parkkinen L, et al. MAPT genetic variation and neuronal maturity alter isoform expression affecting axonal transport in iPSC-derived dopamine neurons. Stem Cell Rep. 2017. https://doi.org/10.1016/j.stemcr.2017.06.005.
Ishikawa T, Imamura K, Kondo T, Koshiba Y, Hara S, Ichinose H, et al. Genetic and pharmacological correction of aberrant dopamine synthesis using patient iPSCs with BH4 metabolism disorders. Human Mol Genet. 2016. https://doi.org/10.1093/hmg/ddw339.
Awad O, Panicker LM, Deranieh RM, Srikanth MP, Brown RA, Voit A, et al. Altered differentiation potential of Gaucher’s disease iPSC neuronal progenitors due to Wnt/β-catenin downregulation. Stem Cell Rep. 2017. https://doi.org/10.1016/j.stemcr.2017.10.029.
Sheng Y, Filichia E, Shick E, Preston KL, Phillips KA, Cooperman L, et al. Using iPSC-derived human DA neurons from opioid-dependent subjects to study dopamine dynamics. Brain Behav. 2016. https://doi.org/10.1002/brb3.491.
Sundberg M, Bogetofte H, Lawson T, Jansson J, Smith G, Astradsson A, et al. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells. 2013. https://doi.org/10.1002/stem.1415.
Fernandes HJR, Patikas N, Foskolou S, Field SF, Park JE, Byrne ML, et al. Single-cell transcriptomics of Parkinson’s disease human in vitro models reveals dopamine neuron-specific stress responses. Cell Rep. 2020. https://doi.org/10.1016/j.celrep.2020.108263.
la Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A. et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016; https://doi.org/10.1016/j.cell.2016.09.027.
Hoffman GE, Hartley BJ, Flaherty E, Ladran I, Gochman P, Ruderfer DM. et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat Commun. 2017;8:2225 https://doi.org/10.1038/s41467-017-02330-5.
Espeso-Gil S, Halene T, Bendl J, Kassim B, ben Hutta G, Iskhakova M, et al. A chromosomal connectome for psychiatric and metabolic risk variants in adult dopaminergic neurons. Genome Med. 2020. https://doi.org/10.1186/s13073-020-0715-x.
Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protocols. 2006. https://doi.org/10.1038/nprot.2006.37.
Yang N, Chanda S, Marro S, Ng YH, Janas JA, Haag D, et al. Generation of pure GABAergic neurons by transcription factor programming. Nat Methods. 2017. https://doi.org/10.1038/nmeth.4291.
Yang N, Chanda S, Südhof T, Wernig M. Generation of pure GABAergic neurons by transcription factor programming. Protocol Exchange. 2017. https://doi.org/10.1038/protex.2017.042.
Miskinyte Id G, Hansen MG, Monni E, Lam M, Bengzon J, Lindvall O, et al. Transcription factor programming of human ES cells generates functional neurons expressing both upper and deep layer cortical markers. PLoS One. 2018. https://doi.org/10.1371/journal.pone.0204688.
Ho S-M, Hartley BJ, TCW J, Beaumont, Stafford M, Slesinger K, et al. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells HHS public access. Methods. 2016;101:113–24. https://doi.org/10.1016/j.ymeth.2015.11.019.
Zhang Y, Pak CH, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013. https://doi.org/10.1016/j.neuron.2013.05.029.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protocols. 2008. https://doi.org/10.1038/nprot.2008.73.
Barretto N, Zhang H, Powell SK, Fernando MB, Zhang S, et al. ASCL1-and DLX2-induced GABAergic neurons from hiPSC-derived NPCs. J Neurosci Methods. 2020. https://doi.org/10.1016/j.jneumeth.2019.108548.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013. https://doi.org/10.1093/bioinformatics/bts635.
Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014. https://doi.org/10.1093/bioinformatics/btt656.
Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019. https://doi.org/10.1093/nar/gkz114.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015. https://doi.org/10.1093/nar/gkv007.
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012. https://doi.org/10.1093/nar/gks042.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009. https://doi.org/10.1093/bioinformatics/btp616.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010. https://doi.org/10.1186/gb-2010-11-3-r25.
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004. https://doi.org/10.2202/1544-6115.1027.
Agarwal D, Sandor C, Volpato V, Caffrey TM, Monzón-Sandoval J, Bowden R, et al. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun. 2020. https://doi.org/10.1038/s41467-020-17876-0.
Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50:621–9. https://doi.org/10.1038/s41588-018-0081-4
Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J Integr Biol. 2012;16:284–7. https://doi.org/10.1089/omi.2011.0118.
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; https://doi.org/10.1093/nar/gkw1092.
Carbon S, Douglass E, Good BM, Unni DR, Harris NL, Mungall CJ, et al. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021. https://doi.org/10.1093/nar/gkaa1113.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000. https://doi.org/10.1038/75556.
Yu G. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol Biol. 2020. https://doi.org/10.1007/978-1-0716-0301-7_11.
Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010. https://doi.org/10.1093/bioinformatics/btq064.
Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012. https://doi.org/10.1093/nar/gks461.
Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, et al. Genetic identification of brain cell types underlying schizophrenia. Nat Genet. 2018. https://doi.org/10.1038/s41588-018-0129-5.
Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019. https://doi.org/10.1038/s41588-018-0269-7.
Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am J Psychiatry. 2017. https://doi.org/10.1176/appi.ajp.2017.16121402.
Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019. https://doi.org/10.1038/s41588-019-0344-8.
Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018. https://doi.org/10.1038/s41593-018-0275-1.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of over 40,000 bipolar disorder cases provides novel biological insights. medRxiv. 2020. https://doi.org/10.1101/2020.09.17.20187054.
Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020. https://doi.org/10.1016/S2215-0366(20)30339-4.
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019. https://doi.org/10.1038/s41593-018-0326-7.
Arnold PD, Askland KD, Barlassina C, Bellodi L, Bienvenu OJ, Black D, et al. Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol Psychiatry. 2018. https://doi.org/10.1038/mp.2017.154.
Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019. https://doi.org/10.1038/s41467-019-12576-w.
Ripke S, Walters JTR, O’Donovan MC. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv. 2020 https://doi.org/10.1101/2020.09.12.20192922.
Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J.Cross-Disorder Group of the Psychiatric Genomics Consortium. et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–1482.e11. https://doi.org/10.1016/j.cell.2019.11.020.
Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiatry. 2018. https://doi.org/10.1038/s41398-018-0150-6.
Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019. https://doi.org/10.1016/S1474-4422(19)30320-5.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015. https://doi.org/10.1371/journal.pcbi.1004219.
Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci USA. 2015. https://doi.org/10.1073/pnas.1504393112.
Rifkin RA, Moss SJ, Slesinger PA. G protein-gated potassium channels: a link to drug addiction. Trends Pharmacol Sci. 2017. https://doi.org/10.1016/j.tips.2017.01.007.
Beckstead, MJ, Grandy DK, Wickman K, Williams JT. Vesicular dopamine release elicits an inhibitory postsynaptic current in midbrain dopamine neurons. Neuron. 2004. https://doi.org/10.1016/j.neuron.2004.05.019.
Cotterill E, Hall D, Wallace K, Mundy WR, Eglen SJ, Shafer TJ. Characterization of early cortical neural network development in multiwell microelectrode array plates. J Biomol Screen. 2016. https://doi.org/10.1177/1087057116640520.
Nedergaard S. A Ca2+-independent slow afterhyperpolarization in substantia nigra compacta neurons. Neuroscience. 2004. https://doi.org/10.1016/j.neuroscience.2004.02.030.
Nedergaard S. Regulation of action potential size and excitability in substantia nigra compacta neurons: Sensitivity to 4-aminopyridine. J Neurophysiol. 1999. https://doi.org/10.1152/jn.1999.82.6.2903.
Bean BP. The action potential in mammalian central neurons. Nat Rev Neurosci. 2007. https://doi.org/10.1038/nrn2148.
Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry All Discip. 2014;55:1068-87. https://doi.org/10.1111/jcpp.12295.
Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardiñas AF, et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet. 2019. https://doi.org/10.1038/s41588-019-0364-4.
Schrode N, Ho S-M, Yamamuro K, Dobbyn A, Huckins L, Matos MR. et al. Synergistic effects of common schizophrenia risk variants. Nat Genet. 2019;51:1475–85. https://doi.org/10.1038/s41588-019-0497-5.
Wray NR, Wijmenga C, Sullivan PF, Yang J, Visscher PM. Common disease is more complex than implied by the core gene omnigenic model. Cell. 2018. https://doi.org/10.1016/j.cell.2018.05.051.
Polimanti R, Walters RK, Johnson EC, McClintick JN, Adkins AE, Adkins DE, et al. Leveraging genome-wide data to investigate differences between opioid use vs. opioid dependence in 41,176 individuals from the Psychiatric Genomics Consortium. Mol Psychiatry. 2020. https://doi.org/10.1038/s41380-020-0677-9.
de Jonge JC, Vinkers CH, Hulshoff Pol HE, Marsman A. GABAergic mechanisms in schizophrenia: linking postmortem and In vivo studies. Front Psychiatry. 2017. https://doi.org/10.3389/fpsyt.2017.00118.
Hauberg ME, Creus-Muncunill J, Bendl J, Kozlenkov A, Zeng B, Corwin C, et al. Common schizophrenia risk variants are enriched in open chromatin regions of human glutamatergic neurons. Nat Commun. 2020. https://doi.org/10.1038/s41467-020-19319-2.
Ragland JD, Maddock RJ, Hurtado MY, Tanase C, Lesh TA, Niendam TA. et al. Disrupted GABAergic facilitation of working memory performance in people with schizophrenia. NeuroImage Clin. 2019;25:102127 https://doi.org/10.1016/j.nicl.2019.102127.
Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018. https://doi.org/10.1126/science.aap8757.
Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013. https://doi.org/10.1038/ng.2711.
Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Pavlides JMW, et al. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018. https://doi.org/10.1016/j.cell.2018.05.046.
Chu HY, Zhen X. Hyperpolarization-activated, cyclic nucleotide-gated (HCN) channels in the regulation of midbrain dopamine systems. Acta Pharmacol Sin. 2010. https://doi.org/10.1038/aps.2010.105.
Picken Bahrey, HL Moody, WJ. Early development of voltage-gated ion currents and firing properties in neurons of the mouse cerebral cortex. Journal of Neurophysiology. 2003. https://doi.org/10.1152/jn.00972.2002.
Rosa F, Dhingra, A, Uysal B, Mendis GDC, Loeffler H, Elsen G, et al. In Vitro Differentiated Human Stem Cell-Derived Neurons Reproduce Synaptic Synchronicity Arising during Neurodevelopment. Stem Cell Reports. 2020. https://doi.org/10.1016/j.stemcr.2020.05.015.
Moore AR, Filipovic R, Mo Z, Rasband MN, Zecevic N, Antic SD. Electrical excitability of early neurons in the human cerebral cortex during the second trimester of gestation. Cerebral Cortex. 2009. https://doi.org/10.1093/cercor/bhn206.
Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015. https://doi.org/10.1038/mp.2014.22.
Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, et al. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018. https://doi.org/10.1016/j.celrep.2018.04.066.
Powell SK, Gregory J, Akbarian S, Brennand KJ. Application of CRISPR/Cas9 to the study of brain development and neuropsychiatric disease. Mol Cell Neurosci. 2017;82. https://doi.org/10.1016/j.mcn.2017.05.007.
Connor JP, Stjepanović D, le Foll B, Hoch E, Budney AJ, Hall WD. Cannabis use and cannabis use disorder. Nat Rev Dis Primers. 2021;7:1–24. https://doi.org/10.1038/s41572-021-00247-4.
Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL. et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 2018;21:1161–70. https://doi.org/10.1038/s41593-018-0206-1.
Cherlyn SYT, Woon PS, Liu JJ, Ong WY, Tsai GC, Sim K. Genetic association studies of glutamate, GABA and related genes in schizophrenia and bipolar disorder: a decade of advance. Neurosci Biobehav Rev. 2010;34:958–977. https://doi.org/10.1016/j.neubiorev.2010.01.002.
Kim Y, Santos R, Gage FH, Marchetto MC. Molecular mechanisms of bipolar disorder: progress made and future challenges. Front Cel Neurosci. 2017;11. https://doi.org/10.3389/fncel.2017.00030.
Ashok AH, Marques TR, Jauhar S, Nour MM, Goodwin GM, Young AH, et al. The dopamine hypothesis of bipolar affective disorder: the state of the art and implications for treatment. Mol Psychiatry. 2017;22:666–679. https://doi.org/10.1038/mp.2017.16.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019. https://doi.org/10.1038/s41588-019-0397-8.
Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. et al. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018;173:1705–1715.e16. https://doi.org/10.1016/j.cell.2018.05.046.
Powell SK, O’Shea C, Brennand KJ, Akbarian S. Parsing the functional impact of noncoding genetic variants in the brain epigenome. Biol Psychiatry. 2021;89;65–75. https://doi.org/10.1016/j.biopsych.2020.06.033.
Acknowledgements
This research was supported by R01MH106056, U01DA047880, R01DA048279, and 6R56MH101454. Figures in this manuscript were created with Biorender.com. The authors wish to thank Rachel Oren for helpful feedback on an earlier version of this manuscript, Dana Infante for assistance in sample collection, and Dr. Stefano Marenco, Dr. Barbara Lipska, and Dr. Pavan Auluck and their staff in the Human Brain Collection Core at the National Institutes of Health for providing postmortem brain tissues.
Author information
Authors and Affiliations
Contributions
SKP, SA, and KJB conceived of the study. SKP, KT, IP, KD, PS, LMH, SA, and KJP designed experiments. SKP, COS, IP, KD, RE, and SH conducted experiments. MI, TL, and AV performed FANS of post-mortem samples. SKP, COS, MI, TL, and AV prepared RNA-sequencing libraries. SKP, KT, and WL conducted computational and bioinformatic analyses. SKP wrote the paper, with contributions from KT and KD. All authors reviewed the manuscript and approved of it in its final form.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Powell, S.K., O’Shea, C., Townsley, K. et al. Induction of dopaminergic neurons for neuronal subtype-specific modeling of psychiatric disease risk. Mol Psychiatry 28, 1970–1982 (2023). https://doi.org/10.1038/s41380-021-01273-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01273-0
This article is cited by
-
Opportunities and limitations for studying neuropsychiatric disorders using patient-derived induced pluripotent stem cells
Molecular Psychiatry (2023)
-
Modeling gene × environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression
Nature Neuroscience (2022)