Abstract
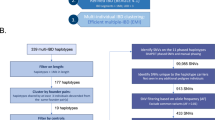
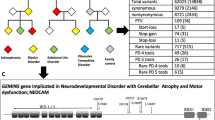
Tourette’s Disorder (TD) is a neurodevelopmental disorder (NDD) that affects about 0.7% of the population and is one of the most heritable NDDs. Nevertheless, because of its polygenic nature and genetic heterogeneity, the genetic etiology of TD is not well understood. In this study, we combined the segregation information in 13 TD multiplex families with high-throughput sequencing and genotyping to identify genes associated with TD. Using whole-exome sequencing and genotyping array data, we identified both small and large genetic variants within the individuals. We then combined multiple types of evidence to prioritize candidate genes for TD, including variant segregation pattern, variant function prediction, candidate gene expression, protein–protein interaction network, candidate genes from previous studies, etc. From the 13 families, 71 strong candidate genes were identified, including both known genes for NDDs and novel genes, such as HtrA Serine Peptidase 3 (HTRA3), Cadherin-Related Family Member 1 (CDHR1), and Zinc Finger DHHC-Type Palmitoyltransferase 17 (ZDHHC17). The candidate genes are enriched in several Gene Ontology categories, such as dynein complex and synaptic membrane. Candidate genes and pathways identified in this study provide biological insight into TD etiology and potential targets for future studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Robertson MM, Eapen V, Singer HS, Martino D, Scharf JM, Paschou P, et al. Gilles de la Tourette syndrome. Nat Rev Dis Prim. 2017;3:16097.
Scharf JM, Miller LL, Gauvin CA, Alabiso J, Mathews CA, Ben-Shlomo Y. Population prevalence of tourette syndrome: a systematic review and meta-analysis. Mov Disord. 2015;30:221–8.
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9.
Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94.
O’Rourke JA, Scharf JM, Platko J, Stewart SE, Illmann C, Geller DA, et al. The familial association of tourette’s disorder and ADHD: the impact of OCD symptoms. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:553–60.
Hirschtritt ME, Lee PC, Pauls DL, Dion Y, Grados MA, Illmann C, et al. Lifetime prevalence, age of risk, and genetic relationships of comorbid psychiatric disorders in Tourette syndrome. JAMA Psychiatry. 2015;72:325–33.
Yu D, Mathews CA, Scharf JM, Neale BM, Davis LK, Gamazon ER, et al. Cross-disorder genome-wide analyses suggest a complex genetic relationship between Tourette’s syndrome and OCD. Am J Psychiatry. 2015;172:82–93.
Davis LK, Yu D, Keenan CL, Gamazon ER, Konkashbaev AI, Derks EM, et al. Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genet. 2013;9:e1003864.
Pringsheim T, Holler-Managan Y, Okun MS. Comprehensive systematic review summary: treatment of tics in people with Tourette syndrome and chronic tic disorders (vol 92, pg 907, 2019). Neurology. 2019;93:415.
Pinto R, Monzani B, Leckman JF, Ruck C, Serlachius E, Lichtenstein P, et al. Understanding the covariation of tics, attention-deficit/hyperactivity, and obsessive-compulsive symptoms: a population-based adult twin study. Am J Med Genet B. 2016;171:938–47.
Darrow SM, Grados M, Sandor P, Hirschtritt ME, Illmann C, Osiecki L, et al. Autism spectrum symptoms in a Tourette’s disorder sample. J Am Acad Child Adolesc Psychiatry. 2017;56:610–7.
Abdulkadir M, Mathews CA, Scharf JM, Yu D, Tischfield JA, Heiman GA, et al. Polygenic risk scores derived from a Tourette syndrome genome-wide association study predict presence of tics in the Avon Longitudinal Study of Parents and Children Cohort. Biol Psychiatry. 2019;85:298–304.
Yu D, Sul JH, Tsetsos F, Nawaz MS, Huang AY, Zelaya I, et al. Interrogating the genetic determinants of Tourette’s syndrome and other tic disorders through genome-wide association studies. Am J Psychiatry. 2019;176:217–27.
Heiman GA, Rispoli J, Seymour C, Leckman JF, King RA, Fernandez TV. Empiric recurrence risk estimates for chronic tic disorders: implications for genetic counseling. Front Neurol. 2020;11:770.
Qi Y, Zheng Y, Li Z, Xiong L. Progress in genetic studies of Tourette’s syndrome. Brain Sci. 2017;7:134.
Deng H, Gao K, Jankovic J. The genetics of Tourette syndrome. Nat Rev Neurol. 2012;8:203–13.
Willsey AJ, Fernandez TV, Yu D, King RA, Dietrich A, Xing J, et al. De novo coding variants are strongly associated with TourettE Disorder. Neuron. 2017;94:486–99.
Wang S, Mandell JD, Kumar Y, Sun N, Morris MT, Arbelaez J, et al. De novo sequence and copy number variants are strongly associated with Tourette disorder and implicate cell polarity in pathogenesis. Cell Rep. 2018;24:3441–54.
Pauls DL, Fernandez TV, Mathews CA, State MW, Scharf JM. The inheritance of Tourette disorder: a review. J Obsessive Compuls Relat Disord. 2014;3:380–5.
Dietrich A, Fernandez TV, King RA, State MW, Tischfield JA, Hoekstra PJ, et al. The Tourette International Collaborative Genetics (TIC Genetics) study, finding the genes causing Tourette syndrome: objectives and methods. Eur Child Adolesc Psychiatry. 2015;24:141–51.
Heiman GA, King RA, Tischfield JA. New Jersey Center for Tourette Syndrome sharing repository: methods and sample description. BMC Med Genomics. 2008;1:58.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders, 4th Edition Text Revision (DSM-IV-TR). Washington DC: American Psychiatric Association; 2000.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders, Fifth Edition (DSM-5). Washington DC: American Psychiatric Association; 2013.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66.
Yandell M, Huff C, Hu H, Singleton M, Moore B, Xing J, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–42.
Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL, et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat Biotechnol. 2014;32:663–9.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13.
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30.
Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199.
Lindsay SJ, Xu YB, Lisgo SN, Harkin LF, Copp AJ, Gerrelli D, et al. HDBR expression: a unique resource for global and individual gene expression studies during early human brain development. Front Neuroanat. 2016;10:86.
Pletscher-Frankild S, Palleja A, Tsafou K, Binder JX, Jensen LJ. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015;74:83–9.
Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8.
Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11:1889–907.
Wong AK, Krishnan A, Troyanskaya OG. GIANT 2.0: genome-scale integrated analysis of gene networks in tissues. Nucleic Acids Res. 2018;46:W65–70.
Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47:569–76.
Huang JK, Carlin DE, Yu MK, Zhang W, Kreisberg JF, Tamayo P, et al. Systematic evaluation of molecular networks for discovery of disease genes. Cell Syst. 2018;6:484–95.
Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–33.
Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34:3572–4.
Sundaram SK, Huq AM, Sun Z, Yu W, Bennett L, Wilson BJ, et al. Exome sequencing of a pedigree with Tourette syndrome or chronic tic disorder. Ann Neurol. 2011;69:901–4.
Moya PR, Wendland JR, Rubenstein LM, Timpano KR, Heiman GA, Tischfield JA, et al. Common and rare alleles of the serotonin transporter gene, SLC6A4, associated with Tourette’s disorder. Mov Disord. 2013;28:1263–70.
Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Stingl K, Mayer AK, Llavona P, Mulahasanovic L, Rudolph G, Jacobson SG, et al. CDHR1 mutations in retinal dystrophies. Sci Rep. 2017;7:6992.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics. 2013;76:7.20.1–41.
Chand V, Nandi D, Mangla AG, Sharma P, Nag A. Tale of a multifaceted co-activator, hADA3: from embryogenesis to cancer and beyond. Open Biol. 2016;6:160153.
Bowden M, Drummond AE, Salamonsen LA, Findlay JK, Nie GY. Evolutionary conservation of mammalian HTRA3 and its developmental regulation in the rat ovary. J Exp Zool B. 2009;312b:701–13.
Naf D, Wilson LA, Bergstrom RA, Smith RS, Goodwin NC, Verkerk A, et al. Mouse models for the Wolf-Hirschhorn deletion syndrome. Hum Mol Genet. 2001;10:91–8.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Yang L, Neale BM, Liu L, Lee SH, Wray NR, Ji N, et al. Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: genome-wide association study of both common and rare variants. Am J Med Genet B. 2013;162b:419–30.
Zhang Q, An Y, Chen ZS, Koon AC, Lau KF, Ngo JCK, et al. A peptidylic inhibitor for neutralizing (r)(GGGGCC)(exp)-associated neurodegeneration in C9ALS-FTD. Mol Ther-Nucl Acids. 2019;16:172–85.
Tsoi H, Lau TCK, Tsang SY, Lau KF, Chan HYE. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc Natl Acad Sci USA. 2012;109:13428–33.
Tsoi H, Chan HYE. Expression of expanded CAG transcripts triggers nucleolar stress in Huntington’s disease. Cerebellum. 2013;12:310–2.
Schabla NM, Mondal K, Swanson PC. DCAF1 (VprBP): emerging physiological roles for a unique dual-service E3 ubiquitin ligase substrate receptor. J Mol Cell Biol. 2019;11:725–35.
Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD. Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene. 2004;23:9230–7.
Kang RJ, Wang L, Sanders SS, Zuo K, Hayden MR, Raymond LA. Altered regulation of striatal neuronal N-methyl-D-aspartate receptor trafficking by palmitoylation in Huntington disease mouse model. Front Synaptic Neurosci. 2019;11:3.
Mahdieh N, Mikaeeli S, Tavasoli AR, Rezaei Z, Maleki M, Rabbani B. Genotype, phenotype and in silico pathogenicity analysis of HEXB mutations: panel based sequencing for differential diagnosis of gangliosidosis. Clin Neurol Neurosurg. 2018;167:43–53.
Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, et al. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175.
Xu C, Aragam N, Li X, Villla EC, Wang L, Briones D, et al. BCL9 and C9orf5 are associated with negative symptoms in schizophrenia: meta-analysis of two genome-wide association studies. PLoS ONE. 2013;8:e51674.
Eicher JD, Powers NR, Miller LL, Akshoomoff N, Amaral DG, Bloss CS, et al. Genome-wide association study of shared components of reading disability and language impairment. Genes Brain Behav. 2013;12:792–801.
Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309.
Acknowledgements
We thank the families who have participated in and contributed to this study. We are also grateful to the NJCTS for facilitating the inception and organization of the TIC Genetics study. This study was supported by a grant from the National Institute of Mental Health (R01MH092293 to GAH and JAT) and by a grant from the New Jersey Center for Tourette Syndrome (to GAH and JAT). This study was also supported by grants from the National Institute of Mental Health (K08MH099424 to TVF) and the National Institute for Environmental Health Science (R01 ES021462 for YSK and BLL). PM has received grants from the Instituto de Salud Carlos III (PI10/01674, PI13/01461), the Consejería de Economía, Innovación, Ciencia y Empresa de la Junta de Andalucía (CVI-02526, CTS-7685), the Consejería de Salud y Bienestar Social de la Junta de Andalucía (PI-0741/2010, PI-0437-2012, PI-0471-2013), the Sociedad Andaluza de Neurología, the Fundación Alicia Koplowitz, the Fundación Mutua Madrileña, and the Jaques and Gloria Gossweiler Foundation. AM has received grants from the Fundacion Alicia Koplowitz and belongs to the research group of the Comissionat per Universitats i Recerca del Departmanent d’Innovacio (DIUE) 2009SGR1119. AM has received grants from the Deutsche Forschungsgemeinschaft (DFG: MU 1692/3-1, MU 1692/4-1, and FOR 2698). AJW received a Young Investigator Award from Tourette Association of America. IH declares that all research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Tourette International Collaborative Genetics Study (TIC Genetics)
Mohamed Abdulkadir1,2,3, Lawrence W. Brown12, Xiaolong Cao1, Barbara J. Coffey13, Li Deng1,2, Andrea Dietrich3, Thomas V. Fernandez4,5, Blanca Garcia-Delgar6, Donald L. Gilbert14, Julie Hagstrøm7, Tammy Hedderly15, Gary A. Heiman1,2, Isobel Heyman16, Pieter J. Hoekstra3, Chaim Huyser17, Eunjoo Kim18, Young-Shin Kim19, Robert A. King4, Justin Koesterich1,2, Yun-Joo Koh20, Samuel Kuperman8, Bennett L. Leventhal21, Marcos Madruga-Garrido22, Athanasios Maras23, Pablo Mir24,25, Astrid Morer6,9,10, Alexander Münchau26, Cara Nasello1,2, Kerstin J. Plessen7,11, Veit Roessner27, Dong-Ho Song28, Matthew W. State13, Joshua K. Thackray1,2, Jay A. Tischfield1,2, A. Jeremy Willsey13,29, Jinchuan Xing1,2, Yeting Zhang1,2, Lisheng Zhou1, Samuel H. Zinner30
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
BJC received research support from NIMH, Emalex, and Teva/Nuvelution; honoraria from American Academy of Child and Adolescent Psychiatry, Harvard Medical School/Psychiatry Academy, and Partners Healthcare. BJC served on the advisory board of Skyland Trail and Teva/Nuvelution, and the following roles at the Tourette Association of America: Co-Chair, Medical Advisory Board; TAA-CDC Partnership. VR has served as advisor or consultancy role for Lilly, Novartis, and Shire, and received conference attendance support or was paid for public speaking by Actelion, Lilly, Medice, Novartis and Shire.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Members of the Tourette International Collaborative Genetics Study (TIC Genetics) are listed below Acknowledgements.
Supplementary information
Rights and permissions
About this article
Cite this article
Cao, X., Zhang, Y., Abdulkadir, M. et al. Whole-exome sequencing identifies genes associated with Tourette’s disorder in multiplex families. Mol Psychiatry 26, 6937–6951 (2021). https://doi.org/10.1038/s41380-021-01094-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01094-1