Abstract
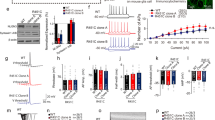
Genetic abnormalities in synaptic proteins are common in individuals with autism; however, our understanding of the cellular and molecular mechanisms disrupted by these abnormalities is limited. SHANK3 is a postsynaptic scaffolding protein of excitatory synapses that has been found mutated or deleted in most patients with 22q13 deletion syndrome and about 2% of individuals with idiopathic autism and intellectual disability. Here, we generated CRISPR/Cas9-engineered human pluripotent stem cells (PSCs) with complete hemizygous SHANK3 deletion (SHANK3 +/–), which is the most common genetic abnormality in patients, and investigated the synaptic and morphological properties of SHANK3-deficient PSC-derived cortical neurons engrafted in the mouse prefrontal cortex. We show that human PSC-derived neurons integrate into the mouse cortex by acquiring appropriate cortical layer identities and by receiving and sending anatomical projections from/to multiple different brain regions. We also demonstrate that SHANK3-deficient human neurons have reduced AMPA-, but not NMDA- or GABA-mediated synaptic transmission and exhibit impaired dendritic arbors and spines, as compared to isogenic control neurons co-engrafted in the same brain region. Together, this study reveals specific synaptic and morphological deficits caused by SHANK3 hemizygosity in human cortical neurons at different developmental stages under physiological conditions and validates the use of co-engrafted control and mutant human neurons as a new platform for studying connectivity deficits in genetic neurodevelopmental disorders associated with autism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16:551–63.
Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4:a009886.
Khan S, Gramfort A, Shetty NR, Kitzbichler MG, Ganesan S, Moran JM, et al. Local and long-range functional connectivity is reduced in concert in autism spectrum disorders. Proc Natl Acad Sci. 2013. https://doi.org/10.1073/pnas.1214533110.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014. https://doi.org/10.1038/nature13772.
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020. https://doi.org/10.1016/j.cell.2019.12.036.
Boeckers TM, Inter C, Smalla KH, Kreutz MR, Bockmann J, Seidenbecher C, et al. Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem Biophys Res Commun. 1999. https://doi.org/10.1006/bbrc.1999.1489.
Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–82.
Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, et al. Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10:e1004580.
Phelan MC, Rogers RC, Saul RA, Stapleton GA, Sweet K, McDermid H, et al. 22q13 deletion syndrome. Am J Med Genet. 2001. https://doi.org/10.1002/1096-8628(20010615)101:2<91::aid-ajmg1340>3.0.co;2-c.
Mitz AR, Philyaw TJ, Boccuto L, Shcheglovitov A, Sarasua SM, Kaufmann WE, et al. Identification of 22q13 genes most likely to contribute to Phelan McDermid syndrome. Eur J Hum Genet. 2018;26:293–302.
Wilson HL, Wong ACC, Shaw SR, Tse W-Y, Stapleton GA, Phelan MC, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–84.
Wang X, Bey AL, Katz BM, Badea A, Kim N, David LK, et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat Commun. 2016. https://doi.org/10.1038/ncomms11459.
Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, Gupta N, et al. Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J Neurosci. 2013;33:18448–68.
Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 2015. https://doi.org/10.1016/j.celrep.2015.04.064.
Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, et al. Importance of Shank3 protein in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. J Biol Chem. 2011;286:34839–50.
Lee J, Chung C, Ha S, Lee D, Kim D-Y, Kim H, et al. Shank3-mutant mice lacking exon 9 show altered excitation/inhibition balance, enhanced rearing, and spatial memory deficit. Front Cell Neurosci. 2015;9:94.
Peixoto RT, Chantranupong L, Hakim R, Levasseur J, Wang W, Merchant T, et al. Abnormal striatal development underlies the early onset of behavioral deficits in Shank3B−/− mice. Cell Rep. 2019. https://doi.org/10.1016/j.celrep.2019.10.021.
Peixoto RT, Wang W, Croney DM, Kozorovitskiy Y, Sabatini BL. Early hyperactivity and precocious maturation of corticostriatal circuits in Shank3B(–/–) mice. Nat Neurosci. 2016. https://doi.org/10.1038/nn.4260.
Speed HE, Kouser M, Xuan Z, Reimers JM, Ochoa CF, Gupta N, et al. Autism-associated insertion mutation (InsG) of Shank3 Exon 21 causes impaired synaptic transmission and behavioral deficits. J Neurosci. 2015;35:9648–65.
Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–42.
Zhou Y, Sharma J, Ke Q, Landman R, Yuan J, Chen H, et al. Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature. 2019. https://doi.org/10.1038/s41586-019-1278-0.
Mei Y, Monteiro P, Zhou Y, Kim J-A, Gao X, Fu Z, et al. Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature. 2016;530:481–4.
Zhao H, Tu Z, Xu H, Yan S, Yan H, Zheng Y, et al. Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Res. 2017;27:1293–7.
Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–108.
Bey AL, Wang X, Yan H, Kim N, Passman RL, Yang Y, et al. Brain region-specific disruption of Shank3 in mice reveals a dissociation for cortical and striatal circuits in autism-related behaviors. Transl Psychiatry. 2018. https://doi.org/10.1038/s41398-018-0142-6.
Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15.
Jiang Yhui, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27.
Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The cellular and molecular landscapes of the developing human central nervous system. Neuron. 2016;89:248–68.
Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013. https://doi.org/10.1038/nature12618.
Kathuria A, Nowosiad P, Jagasia R, Aigner S, Taylor RD, Andreae LC, et al. Stem cell-derived neurons from autistic individuals with SHANK3 mutation show morphogenetic abnormalities during early development. Mol Psychiatry. 2018. https://doi.org/10.1038/mp.2017.185.
Yi F, Danko T, Botelho SC, Patzke C, Pak C, Wernig M, et al. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science. 2016. https://doi.org/10.1126/science.aaf2669.
Gouder L, Vitrac A, Goubran-Botros H, Danckaert A, Tinevez JY, André-Leroux G, et al. Altered spinogenesis in iPSC-derived cortical neurons from patients with autism carrying de novo SHANK3 mutations. Sci Rep. 2019. https://doi.org/10.1038/s41598-018-36993-x.
Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009. https://doi.org/10.1038/nbt.1529.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013. https://doi.org/10.1038/nprot.2013.143.
Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011. https://doi.org/10.1038/nature09714.
Viswanathan S, Williams ME, Bloss EB, Stasevich TJ, Speer CM, Nern A, et al. High-performance probes for light and electron microscopy. Nat Methods. 2015. https://doi.org/10.1038/nmeth.3365.
Kim EJ, Jacobs MW, Ito-Cole T, Callaway EM. Improved monosynaptic neural circuit tracing using engineered rabies virus glycoproteins. Cell Rep. 2016. https://doi.org/10.1016/j.celrep.2016.03.067.
Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015. https://doi.org/10.1016/j.cell.2015.11.025.
Chiola S, Do MD, Centrone L, Mallamaci A. Foxg1 overexpression in neocortical pyramids stimulates dendrite elongation via Hes1 and pCreb1 upregulation. Cereb Cortex. 2018. https://doi.org/10.1093/cercor/bhy007.
Jin C, Kang HR, Kang H, Zhang Y, Lee Y, Kim Y, et al. Unexpected compensatory increase in Shank3 transcripts in Shank3 knock-out mice having partial deletions of exons. Front Mol Neurosci. 2019. https://doi.org/10.3389/fnmol.2019.00228.
Ching TT, Maunakea AK, Jun P, Hong C, Zardo G, Pinkel D, et al. Epigenome analyses using BAG microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet. 2005. https://doi.org/10.1038/ng1563.
Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, Dsouza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010. https://doi.org/10.1038/nature09165.
Ährlund-Richter S, Xuan Y, van Lunteren JA, Kim H, Ortiz C, Pollak Dorocic I, et al. A whole-brain atlas of monosynaptic input targeting four different cell types in the medial prefrontal cortex of the mouse. Nat Neurosci. 2019. https://doi.org/10.1038/s41593-019-0354-y.
Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011. https://doi.org/10.1038/nature09915.
Sarkar A, Mei A, Paquola ACM, Stern S, Bardy C, Klug JR, et al. Efficient generation of CA3 neurons from human pluripotent stem cells enables modeling of hippocampal connectivity in vitro. Cell Stem Cell. 2018. https://doi.org/10.1016/j.stem.2018.04.009.
Osakada F, Callaway EM. Design and generation of recombinant rabies virus vectors. Nat Protoc. 2013. https://doi.org/10.1038/nprot.2013.094.
Maisano X, Litvina E, Tagliatela S, Aaron GB, Grabel LB, Naegele JR. Differentiation and functional incorporation of embryonic stem cell-derived GABAergic interneurons in the dentate gyrus of mice with temporal lobe epilepsy. J Neurosci. 2012. https://doi.org/10.1523/jneurosci.2683-11.2012.
Cunningham M, Cho JH, Leung A, Savvidis G, Ahn S, Moon M, et al. hPSC-derived maturing GABAergic interneurons ameliorate seizures and abnormal behavior in epileptic mice. Cell Stem Cell. 2014. https://doi.org/10.1016/j.stem.2014.10.006.
Steinbeck JA, Choi SJ, Mrejeru A, Ganat Y, Deisseroth K, Sulzer D, et al. Optogenetics enables functional analysis of human embryonic stem cell-derived grafts in a Parkinson’s disease model. Nat Biotechnol. 2015. https://doi.org/10.1038/nbt.3124.
Adler AF, Cardoso T, Nolbrant S, Mattsson B, Hoban DB, Jarl U, et al. hESC-derived dopaminergic transplants integrate into basal ganglia circuitry in a preclinical model of Parkinson’s disease. Cell Rep. 2019. https://doi.org/10.1016/j.celrep.2019.08.058.
Espuny-Camacho I, Arranz AM, Fiers M, Snellinx A, Ando K, Munck S, et al. Hallmarks of Alzheimer’s disease in stem-cell-derived human neurons transplanted into mouse brain. Neuron. 2017. https://doi.org/10.1016/j.neuron.2017.02.001.
Linaro D, Vermaercke B, Iwata R, Ramaswamy A, Libé-Philippot B, Boubakar L, et al. Xenotransplanted human cortical neurons reveal species-specific development and functional integration into mouse visual circuits. Neuron. 2019;104:972.e6.
Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. XCoexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013. https://doi.org/10.1016/j.cell.2013.10.020.
Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–21.
Koshino H, Kana RK, Keller TA, Cherkassky VL, Minshew NJ, Just MA. fMRI investigation of working memory for faces in autism: visual coding and underconnectivity with frontal areas. Cereb Cortex. 2008. https://doi.org/10.1093/cercor/bhm054.
Weng SJ, Wiggins JL, Peltier SJ, Carrasco M, Risi S, Lord C, et al. Alterations of resting state functional connectivity in the default network in adolescents with autism spectrum disorders. Brain Res. 2010. https://doi.org/10.1016/j.brainres.2009.11.057.
Ibrahim K, Eilbott JA, Ventola P, He G, Pelphrey KA, McCarthy G, et al. Reduced amygdala–prefrontal functional connectivity in children with autism spectrum disorder and co-occurring disruptive behavior. Biol Psychiatry Cogn Neurosci Neuroimaging. 2019. https://doi.org/10.1016/j.bpsc.2019.01.009.
Grealish S, Heuer A, Cardoso T, Kirkeby A, Jönsson M, Johansson J, et al. Monosynaptic tracing using modified rabies virus reveals early and extensive circuit integration of human embryonic stem cell-derived neurons. Stem Cell Reports. 2015. https://doi.org/10.1016/j.stemcr.2015.04.011.
Denardo LA, Berns DS, Deloach K, Luo L. Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nat Neurosci. 2015. https://doi.org/10.1038/nn.4131.
Feng D, Lau C, Ng L, Li Y, Kuan L, Sunkin SM, et al. Exploration and visualization of connectivity in the adult mouse brain. Methods. 2015. https://doi.org/10.1016/j.ymeth.2015.01.009.
Scannevin RH, Huganir RL. Postsynaptic organisation and regulation of excitatory synapses. Nat Rev Neurosci. 2000. https://doi.org/10.1038/35039075.
Nusser Z, Lujan R, Laube G, Roberts JDB, Molnar E, Somogyi P. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 1998. https://doi.org/10.1016/S0896-6273(00)80565-6.
Matsuzaki M, Ellis-Davies GCR, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001. https://doi.org/10.1038/nn736.
Yuste R, Bonhoeffer T. Genesis of dendritic spines: Insights from ultrastructural and imaging studies. Nat Rev Neurosci. 2004;5:24–34.
Li Z, Sheng M. Some assembly required: the development of neuronal synapses. Nat Rev Mol Cell Biol. 2003;4:833–41.
Kolluri N, Sun Z, Sampson AR, Lewis DA Lamina-specific reductions in dendritic spine density in the prefrontal cortex of subjects with schizophrenia. Am J Psychiatry. 2005. https://doi.org/10.1176/appi.ajp.162.6.1200.
Lewis DA, González-Burgos G. Neuroplasticity of neocortical circuits in schizophrenia. Neuropsychopharmacology. 2008;33:141–65.
Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000. https://doi.org/10.1001/archpsyc.57.1.65.
Jacobs B, Schall M, Prather M, Kapler E, Driscoll L, Baca S, et al. Regional dendritic and spine variation in human cerebral cortex: a quantitative golgi study. Cereb Cortex. 2001. https://doi.org/10.1093/cercor/11.6.558.
Petanjek Z, Judaš M, Šimić G, Rašin MR, Uylings HBM, Rakic P, et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci USA. 2011. https://doi.org/10.1073/pnas.1105108108.
Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, et al. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005. https://doi.org/10.1523/JNEUROSCI.4354-04.2005.
Pina-Crespo JC, Talantova M, Cho E-G, Soussou W, Dolatabadi N, Ryan SD, et al. High-frequency hippocampal oscillations activated by optogenetic stimulation of transplanted human ESC-derived neurons. J Neurosci. 2012. https://doi.org/10.1523/jneurosci.3735-12.2012.
Nagashima F, Suzuki IK, Shitamukai A, Sakaguchi H, Iwashita M, Kobayashi T, et al. Novel and robust transplantation reveals the acquisition of polarized processes by cortical cells derived from mouse and human pluripotent stem cells. Stem Cells Dev. 2013. https://doi.org/10.1089/scd.2013.0251.
Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013. https://doi.org/10.1016/j.neuron.2012.12.011.
Oki K, Tatarishvili J, Wood J, Koch P, Wattananit S, Mine Y, et al. Human-induced pluripotent stem cells form functional neurons and improve recovery after grafting in stroke-damaged brain. Stem Cells. 2012. https://doi.org/10.1002/stem.1104.
Vicidomini C, Ponzoni L, Lim D, Schmeisser M, Reim D, Morello N, et al. Pharmacological enhancement of mGlu5 receptors rescues behavioral deficits in SHANK3 knock-out mice. Mol Psychiatry. 2017;22:689–702.
Acknowledgements
We thank Ricardo Dolmetsch and Theo Palmer for providing iPSC lines; Yuanyuan Wu for helping with generating homozygous CRISPR/Cas9-engineered stem cells; Travis Philyaw for helping with mapping deletions; Brittney Nhem and Nico Edgar for helping with image analysis; Megan Williams, Edward Callaway, Liqun Luo, and John Naughton for plasmids; Chris Rodesch and the University of Utah Cell Imaging Core for assisting with microscopy, Jay Spampanato and Peter West for advising on intracranial injections and slice physiology; Monica Vetter, Richard Dorsky, and Sungjin Park for commenting on the manuscript. This work was supported by grants from the NIMH, NINDS, Brain Research Foundation, Brain and Behavior Research Foundation, Whitehall Foundation, Alan B. Slifka Foundation, and Utah Neuroscience and Genome Project Initiatives.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chiola, S., Napan, K.L., Wang, Y. et al. Defective AMPA-mediated synaptic transmission and morphology in human neurons with hemizygous SHANK3 deletion engrafted in mouse prefrontal cortex. Mol Psychiatry 26, 4670–4686 (2021). https://doi.org/10.1038/s41380-021-01023-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01023-2
This article is cited by
-
Restoration of nNOS Expression Rescues Autistic-Like Phenotypes Through Normalization of AMPA Receptor-Mediated Neurotransmission
Molecular Neurobiology (2024)
-
Haploinsufficiency of Shank3 increases the orientation selectivity of V1 neurons
Scientific Reports (2022)
-
Analyses of the autism-associated neuroligin-3 R451C mutation in human neurons reveal a gain-of-function synaptic mechanism
Molecular Psychiatry (2022)
-
Modeling human telencephalic development and autism-associated SHANK3 deficiency using organoids generated from single neural rosettes
Nature Communications (2022)
-
iPSC toolbox for understanding and repairing disrupted brain circuits in autism
Molecular Psychiatry (2022)