Abstract
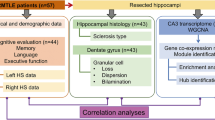
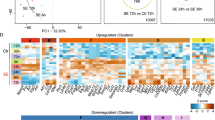
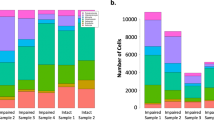
We have previously demonstrated functional and molecular changes in hippocampal subfields in individuals with schizophrenia (SZ) psychosis associated with hippocampal excitability. In this study, we use RNA-seq and assess global transcriptome changes in the hippocampal subfields, DG, CA3, and CA1 from individuals with SZ psychosis and controls to elucidate subfield-relevant molecular changes. We also examine changes in gene expression due to antipsychotic medication in the hippocampal subfields from our SZ ON- and OFF-antipsychotic medication cohort. We identify unique subfield-specific molecular profiles in schizophrenia postmortem samples compared with controls, implicating astrocytes in DG, immune mechanisms in CA3, and synaptic scaling in CA1. We show a unique pattern of subfield-specific effects by antipsychotic medication on gene expression levels with scant overlap of genes differentially expressed by SZ disease effect versus medication effect. These hippocampal subfield changes serve to confirm and extend our previous model of SZ and can explain the lack of full efficacy of conventional antipsychotic medication on SZ symptomatology. With future characterization using single-cell studies, the identified distinct molecular profiles of the DG, CA3, and CA1 in SZ psychosis may serve to identify further potential hippocampal-based therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
Custom R codes and data to support the data analysis are available at https://github.com/konopkalab/Hippo_Subfields.
References
Andreasen B. Introductory textbook of psychiatry. 4th ed. American Psychiatric, Washington, DC, 2006.
Vos T, Allen C, Arora M, Barber RM, Bhutta ZA, Brown A, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–602.
Strauss JS, Carpenter WT, Bartko JJ. The diagnosis and understanding of schizophrenia. Part III. Speculations on the processes that underlie schizophrenic symptoms and signs. Schizophr Bull. 1974;11:61–9.
Davis JM, Casper R. Antipsychotic drugs. Drugs. 1977;14:260–82.
Stone JM, Raffin M, Morrison P, Mcguire PK. The biological basis of antipsychotic response in schizophrenia. J Psychopharmacol. 2010;24:953–64.
Conley RR, Kelly DL. Management of treatment resistance in schizophrenia. Biol Psychiatry. 2001;50:898–911.
Uçok A, Gaebel W. Side effects of atypical antipsychotics: a brief overview. World Psychiatry. 2008;7:58–62.
Valenstein M, Blow FC, Copeland LA, McCarthy JF, Zeber JE, Gillon L, et al. Poor antipsychotic adherence among patients with schizophrenia: medication and patient factors. Schizophr Bull. 2004;30:255–64.
Heckers S, Stone D, Walsh J, Shick J, Koul P, Benes FM. Differential hippocampal expression of glutamic acid decarboxylase 65 and 67 messenger RNA in bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2002;59:521–29.
Sinkus ML, Adams CE, Logel J, Freedman R, Leonard S. Expression of immune genes on chromosome 6p21.3-22.1 in schizophrenia. Brain Behav Immun. 2013;32:51–62.
Benes FM. Evidence for altered trisynaptic circuitry in schizophrenic hippocampus. In: Biological psychiatry. Biological Psychiatry, 1999;46:589–99.
Ruzicka W, Subburaju S, Benes FM. Circuit- and diagnosis-specific DNA methylation changes at gamma-aminobutyric acid-related genes in postmortem human hippocampus in schizophrenia and bipolar disorder. JAMA Psychiatry. 2015;72:541–51.
Tamminga CA, Stan AD, Wagner AD. The hippocampal formation in schizophrenia. Am J Psychiatry. 2010;167:1178–93.
Li W, Ghose S, Gleason K, Begovic A, Perez J, Bartko J, et al. Synaptic proteins in schizophrenia hippocampus indicate increased neuronal activity in CA3. Am J Psychiatry. 2015;172:373–82.
Segev A, Yanagi M, Scott D, Southcott SA, Lister JM, Tan C, et al. Reduced GluN1 in mouse dentate gyrus is associated with CA3 hyperactivity and psychosis-like behaviors. Mol Psychiatry. 2018. https://doi.org/10.1038/s41380-018-0124-3.
Yassa MA, Stark CE. Pattern separation in the hippocampus. Trends Neurosci. 2011;34:515–25.
Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, et al. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11.
Ghose S, Winter MK, McCarson KE, Tamminga CA, Enna SJ. The GABAβ receptor as a target for antidepressant drug action. Br J Pharmacol. 2011;162:1–17.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–5.
Anders S, Pyl PT, Huber W. HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300.
Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–23.
Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014;13:13–24.
Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8.
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Gao X, Sakai K, Roberts RC, Conley RR, Dean B, Tamminga CA. Ionotropic glutamate receptors and expression of N-Methyl-D-aspartate receptor subunits in subregions of human hippocampus: effects of schizophrenia. Am J Psychiatry. 2000;157:1141–9.
Kolomeets NS, Orlovskaya DD, Rachmanova VI, Uranova NA. Ultrastructural alterations in hippocampal mossy fiber synapses in schizophrenia: a postmortem morphometric study. Synapse. 2005;57:47–55.
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Mexal S, Berger R, Logel J, Ross RG, Freedman R, Leonard S. Differential regulation of α7 nicotinic receptor gene (CHRNA7) expression in schizophrenic smokers. J Mol Neurosci. 2010;40:185–95.
Gouvêa ES, Santos Filho AF, Ota VK, Mrad V, Gadelha A, Bressan RA, et al. The role of the CNR1 gene in schizophrenia: a systematic review including unpublished data. Rev Bras Psiquiatr. 2017;39:160–71.
Kang WS, Park JK, Kim SK, Park HJ, Lee SM, Song JY, et al. Genetic variants of GRIA1 are associated with susceptibility to schizophrenia in Korean population. Mol Biol Rep. 2012;39:10697–703.
Fang J, Wang Y, Lv X, Shen X, Ni X, Ding K. Structure of a β-glucan from Grifola frondosa and its antitumor effect by activating Dectin-1/Syk/NF-κB signaling. Glycoconj J. 2012;29:365–77.
Fric J, Zelante T, Wong AYW, Mertes A, Yu H-B, Ricciardi-Castagnoli P, et al. NFAT control of innate immunity. Blood. 2012;120:1380–9.
Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu Rev Immunol. 2012;30:491–529.
Plato A, Willment JA, Brown GD. C-type lectin-like receptors of the dectin-1 cluster: ligands and signaling pathways. Int Rev Immunol. 2013;32:134–56.
Iwakura Y, Nawa H. ErbB1-4-dependent EGF/neuregulin signals and their cross talk in the central nervous system: pathological implications in schizophrenia and Parkinson’s disease. Front Cell Neurosci. 2013;7:1–13.
Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, Kouassi E. Inflammatory cytokine alterations in schizophrenia: a systematic quantitative review. Biol Psychiatry. 2008;63:801–8.
Fillman SG, Cloonan N, Catts VS, Miller LC, Wong J, McCrossin T, et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2013;18:206–14.
Pasternak O, Kubicki M, Shenton ME. In vivo imaging of neuroinflammation in schizophrenia. Schizophr Res. 2016;173:200–12.
Trépanier MO, Hopperton KE, Mizrahi R, Mechawar N, Bazinet RP. Postmortem evidence of cerebral inflammation in schizophrenia: a systematic review. Mol Psychiatry. 2016;21:1009–26.
van Kesteren CF, Gremmels H, de Witte LD, Hol EM, Van Gool AR, Falkai PG, et al. Immune involvement in the pathogenesis of schizophrenia: a meta-analysis on postmortem brain studies. Transl Psychiatry. 2017;7:1–11.
Hwang Y, Kim J, Shin JY, Kim JII, Seo JS, Webster MJ, et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry. 2013;3:1–9.
Crespo-Facorro B, Prieto C, Sainz J. Schizophrenia gene expression profile reverted to normal levels by antipsychotics. Int J Neuropsychopharmacol. 2015;18:1–7.
Santoro ML, Ota VK, Stilhano RS, Silva PN, Santos CM, Diana MC, et al. Effect of antipsychotic drugs on gene expression in the prefrontal cortex and nucleus accumbens in the spontaneously hypertensive rat (SHR). Schizophr Res. 2014;157:163–8.
Kalmady SV, Agrawal R, Venugopal D, Shivakumar V, Amaresha AC, Agarwal SM, et al. CHRFAM7A gene expression in schizophrenia: clinical correlates and the effect of antipsychotic treatment. J Neural Transm. 2018;125:741–8.
Kaplan MS, Hinds JW. Neurogenesis in the adult rat: electron microscopic analysis of light radioautographs. Science. 1977;197:1092–4.
Maekawa M, Takashima N, Arai Y, Nomura T, Inokuchi K, Yuasi S, et al. Pax6 is required for production and maintenance of progenitor cells in postnatal hippocampal neurogenesis. Genes Cells. 2005;10:1001–14.
Sakurai K, Osumi N. The neurogenesis-controlling factor, pax6, inhibits proliferation and promotes maturation in murine astrocytes. J Neurosci. 2008;28:4604–12.
Klempin F, Marr RA, Peterson DA. Modification of Pax6 and Olig2 expression in adult hippocampal neurogenesis selectively induces stem cell fate and alters both neuronal and glial populations. Stem Cells. 2012;30:500–9.
Dwyer DS, Weeks K, Aamodt EJ. Drug discovery based on genetic and metabolic findings in schizophrenia. Expert Rev Clin Pharmacol. 2008;1:773–89.
Weeks KR, Dwyer DS, Aamodt EJ. Antipsychotic drugs activate the C. elegans AKT pathway via the DAF-2 insulin/IGF-1 receptor. ACS Chem Neurosci. 2010;1:463–73.
Bowling H, Zhang G, Bhattacharya A, Pérez-Cuesta LM, Deinhardt K, Hoeffer CA, et al. Antipsychotics activate mTORC1-dependent translation to enhance neuronal morphological complexity. Sci Signal. 2014;7:4.
Mchugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, et al. Dentate Gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science. 2007;317:94–99.
Tronel S, Belnoue L, Grosjean N, Revest JM, Piazza PV, Koehl M, et al. Adult-born neurons are necessary for extended contextual discrimination. Hippocampus. 2012;22:292–8.
Clelland CD, Choi M, Romberg C, Clemenson GD, Fragniere A, Tyers P, et al. A functional role for adult hippocampal neurogenesis in spatial pattern separation. Science. 2009;325:210–3.
Sommer IE, de Witte L, Begemann M, Kahn RS. Nonsteroidal anti-inflammatory drugs in schizophrenia. J Clin Psychiatry. 2012;73:414–9.
Nitta M, Kishimoto T, Müller N, Weiser M, Davidson M, Kane JM, et al. Adjunctive use of nonsteroidal anti-inflammatory drugs for schizophrenia: a meta-analytic investigation of randomized controlled trials. Schizophr Bull. 2013;39:1230–41.
Konstantinopoulos PA, Lehmann DF. The cardiovascular toxicity of selective and nonselective cyclooxygenase inhibitors: Comparisons, contrasts, and aspirin confounding. J Clin Pharmacol. 2005;45:742–50.
Lyketsos CG, Breitner JCS, Green RC, Martin BK, Meinert C, Piantadosi S, et al. Naproxen and celecoxib do not prevent AD in early results from a randomized controlled trial. Neurology. 2007;68:1800–8.
Martin BK, Szekely C, Brandt J, Piantadosi S, Breitner JCS, Craft S, et al. Cognitive function over time in the Alzheimer’s disease anti-inflammatory prevention trial (ADAPT): Results of a randomized, controlled trial of naproxen and celecoxib. Arch Neurol. 2008;65:896–905.
Cimino PJ, Sokal I, Leverenz J, Fukui Y, Montine TJ. DOCK2 is a microglial specific regulator of central nervous system innate immunity found in normal and Alzheimer’s disease brain. Am J Pathol. 2009;175:1622–30.
Dissing-Olesen L, LeDue JM, Rungta RL, Hefendehl JK, Choi HB, MacVicar BA. Activation of neuronal NMDA receptors triggers transient ATP-mediated microglial process outgrowth. J Neurosci. 2014;34:10511–27.
Eyo UB, Peng J, Swiatkowski P, Mukherjee A, Bispo A, Wu LJ. Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus. J Neurosci. 2014;34:10528–40.
Wu Y, Dissing-Olesen L, MacVicar BA, Stevens B. Microglia: dynamic mediators of synapse development and plasticity. Trends Immunol. 2015;36:605–13.
Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, Lafaille JJ, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–609.
Yao I, Iida J, Nishimura W, Hata Y. Synaptic localization of SAPAP1, a synaptic membrane-associated protein. Genes Cells. 2003;8:121–9.
Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DT, Sheng M, et al. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci. 2012;15:1655–66.
Kohen R, Dobra A, Tracy JH, Haugen E. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl Psychiatry. 2014;4:e366.
Collado-Torres L, Burke EE, Peterson A, Shin JH, Straub RE, Rajpurohit A, et al. Regional heterogeneity in gene expression, regulation, and coherence in the frontal cortex and hippocampus across development and schizophrenia. Neuron. 2019;103:203–e8.
Bobilev AM, Perez JM, Tamminga CA. Molecular alterations in the medial temporal lobe in schizophrenia. Schizophr Res. 2019. https://doi.org/10.1016/j.schres.2019.06.001.
Acknowledgements
We wish to thank the next of kin of the brain tissue donors who made this study possible, the Dallas County Medical Examiners’ Office, UT Southwestern Transplant Service and Willed Body Program for assistance with procurement of tissue. This project could not have been performed without the support and generosity of Dr Kenneth Altshuler. This project was performed with funding from the Stanton Sharp Distinguished Chair Endowment and T32 MH076690 Basic Science Training Program in Neurobiology of Mental Illness.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Perez, J.M., Berto, S., Gleason, K. et al. Hippocampal subfield transcriptome analysis in schizophrenia psychosis. Mol Psychiatry 26, 2577–2589 (2021). https://doi.org/10.1038/s41380-020-0696-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-0696-6
This article is cited by
-
Cell type specific transcriptomic differences in depression show similar patterns between males and females but implicate distinct cell types and genes
Nature Communications (2023)
-
Distinct contributions of GluA1-containing AMPA receptors of different hippocampal subfields to salience processing, memory and impulse control
Translational Psychiatry (2022)
-
Hippocampal volume and hippocampal neuron density, number and size in schizophrenia: a systematic review and meta-analysis of postmortem studies
Molecular Psychiatry (2021)
-
Linking proteomic alterations in schizophrenia hippocampus to NMDAr hypofunction in human neurons and oligodendrocytes
European Archives of Psychiatry and Clinical Neuroscience (2021)
-
In silico hippocampal modeling for multi-target pharmacotherapy in schizophrenia
npj Schizophrenia (2020)