Abstract
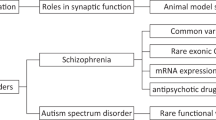
Mutations in the family of neurexins (NRXN1, NRXN2 and NRXN3) have been repeatedly identified in patients with autism spectrum disorder (ASD) and schizophrenia (SCZ). However, it remains unclear how these DNA variants affect neurexin functions and thereby predispose to these neurodevelopmental disorders. Understanding both the wild-type and pathologic roles of these genes in the brain could help unveil biological mechanisms underlying mental disorders. In this regard, numerous studies have focused on generating relevant loss-of-function (LOF) mammalian models. Although this has increased our knowledge about their normal functions, the potential pathologic role(s) of these human variants remains elusive. Indeed, after reviewing the literature, it seems apparent that a traditional LOF-genetic approach based on complete LOF might not be sufficient to unveil the role of these human mutations. First, these genes present a very complex transcriptome and total-LOF of all isoforms may not be the cause of toxicity in patients, particularly given evidence that causative variants act through haploinsufficiency. Moreover, human DNA variants may not all lead to LOF but potentially to intricate transcriptome changes that could also include the generation of aberrant isoforms acting as a gain-of-function (GOF). Furthermore, their transcriptomic complexity most likely renders them prone to genetic compensation when one tries to manipulate them using traditional site-directed mutagenesis approaches, and this could act differently from model to model leading to heterogeneous and conflicting phenotypes. This review compiles the relevant literature on variants identified in human studies and on the mouse models currently deployed, and offers suggestions for future research.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bailey A, Lecouteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, et al. Autism as a strongly genetic disorder - evidence from a British Twin Study. Psychol Med. 1995;25:63–77.
Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–92.
De Crescenzo F, Postorino V, Siracusano M, Riccioni A, Armando M, Curatolo P, et al. Autistic symptoms in schizophrenia spectrum disorders: a systematic review and meta-analysis. Front Psychiatry. 2019;10:78.
Lord C, Elsabbagh M, Baird G, Veenstra-Vanderweele J. Autism spectrum disorder. Lancet. 2018;392:508–20.
Asarnow RF, Forsyth JK. Genetics of childhood-onset schizophrenia. Child Adolesc Psychiatr Clin N Am. 2013;22:675–87.
Ahn K, An SS, Shugart YY, Rapoport JL. Common polygenic variation and risk for childhood-onset schizophrenia. Mol Psychiatry. 2016;21:94–96.
Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nurnberger JI Jr., Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet. 2017;18:362–76.
Foley C, Corvin A, Nakagome S. Genetics of schizophrenia: ready to translate? Curr Psychiatry Rep. 2017;19:61.
Schaaf CP, Wiszniewska J, Beaudet AL. Copy number and SNP arrays in clinical diagnostics. Annu Rev Genomics Hum Genet. 2011;12:25–51.
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–7.
Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35.
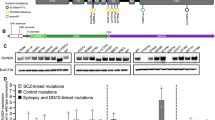
Bena F, Bruno DL, Eriksson M, van Ravenswaaij-Arts C, Stark Z, Dijkhuizen T, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:388–403.
Lowther C, Speevak M, Armour CM, Goh ES, Graham GE, Li C, et al. Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genet Med. 2017;19:53–61.
Sudhof TC. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell. 2017;171:745–69.
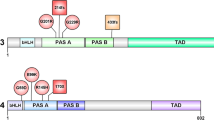
Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507.
Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56.
Tabuchi K, Sudhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–59.
Missler M, Sudhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26.
Treutlein B, Gokce O, Quake SR, Sudhof TC. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA. 2014;111:e1291–99.
Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13.
Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–28.
Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207.
Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–56.
Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, Pugh T, et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–43.
Duong L, Klitten LL, Moller RS, Ingason A, Jakobsen KD, Skjodt C, et al. Mutations in NRXN1 in a family multiply affected with brain disorders: NRXN1 mutations and brain disorders. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:354–8.
Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011;130:563–73.
Mohrmann I, Gillessen-Kaesbach G, Siebert R, Caliebe A, Hellenbroich Y. A de novo 0.57 Mb microdeletion in chromosome 11q13.1 in a patient with speech problems, autistic traits, dysmorphic features and multiple endocrine neoplasia type 1. Eur J Med Genet. 2011;54:e461–464.
Boyle MI, Jespersgaard C, Nazaryan L, Ravn K, Brondum-Nielsen K, Bisgaard AM, et al. Deletion of 11q12.3-11q13.1 in a patient with intellectual disability and childhood facial features resembling Cornelia de Lange syndrome. Gene. 2015;572:130–4.
Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, et al. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet. 2012;90:133–41.
Yuan H, Wang Q, Liu Y, Yang W, He Y, Gusella JF, et al. A rare exonic NRXN3 deletion segregating with neurodevelopmental and neuropsychiatric conditions in a three-generation Chinese family. Am J Med Genet B Neuropsychiatr Genet. 2018;177:589–95.
Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–65.
Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43.
Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Genetic R. Outcome in Psychosis C et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–10.
Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–96.
Hu Z, Xiao X, Zhang Z, Li M. Genetic insights and neurobiological implications from NRXN1 in neuropsychiatric disorders. Mol Psychiatry. 2019;24:1400–14.
Todarello G, Feng N, Kolachana BS, Li C, Vakkalanka R, Bertolino A, et al. Incomplete penetrance of NRXN1 deletions in families with schizophrenia. Schizophr Res. 2014;155:1–7.
Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373.
Levinson DF, Shi J, Wang K, Oh S, Riley B, Pulver AE, et al. Genome-wide association study of multiplex schizophrenia pedigrees. Am J Psychiatry. 2012;169:963–73.
Ikeda M, Aleksic B, Kirov G, Kinoshita Y, Yamanouchi Y, Kitajima T, et al. Copy number variation in schizophrenia in the Japanese population. Biol Psychiatry. 2010;67:283–6.
Li Z, Chen J, Xu Y, Yi Q, Ji W, Wang P, et al. Genome-wide analysis of the role of copy number variation in schizophrenia risk in Chinese. Biol Psychiatry. 2016;80:331–7.
Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204:108–14.
Szatkiewicz JP, O’Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–73.
Morris DW, Pearson RD, Cormican P, Kenny EM, O’Dushlaine CT, Perreault LP, et al. An inherited duplication at the gene p21 Protein-Activated Kinase 7 (PAK7) is a risk factor for psychosis. Hum Mol Genet. 2014;23:3316–26.
Guha S, Rees E, Darvasi A, Ivanov D, Ikeda M, Bergen SE, et al. Implication of a rare deletion at distal 16p11.2 in schizophrenia. JAMA Psychiatry. 2013;70:253–60.
Buizer-Voskamp JE, Muntjewerff JW, Genetic R, Outcome in Psychosis Consortium M, Strengman E, Sabatti C, et al. Genome-wide analysis shows increased frequency of copy number variation deletions in Dutch schizophrenia patients. Biol Psychiatry. 2011;70:655–62.
International Schizophrenia C. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41.
Magri C, Sacchetti E, Traversa M, Valsecchi P, Gardella R, Bonvicini C, et al. New copy number variations in schizophrenia. PLoS One. 2010;5:e13422.
Costain G, Lionel AC, Merico D, Forsythe P, Russell K, Lowther C, et al. Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet. 2013;22:4485–501.
Rudd DS, Axelsen M, Epping EA, Andreasen NC, Wassink TH. A genome-wide CNV analysis of schizophrenia reveals a potential role for a multiple-hit model. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:619–26.
Priebe L, Degenhardt F, Strohmaier J, Breuer R, Herms S, Witt SH, et al. Copy number variants in German patients with schizophrenia. PLoS One. 2013;8:e64035.
Van Den Bossche MJ, Johnstone M, Strazisar M, Pickard BS, Goossens D, Lenaerts AS, et al. Rare copy number variants in neuropsychiatric disorders: specific phenotype or not? Am J Med Genet B Neuropsychiatr Genet. 2012;159B:812–22.
Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6.
Ahn K, Gotay N, Andersen TM, Anvari AA, Gochman P, Lee Y, et al. High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry. 2014;19:568–72.
Hu XF, Zhang JS, Jin C, Mi WF, Wang F, Ma WB, et al. Association study of NRXN3 polymorphisms with schizophrenia and risperidone-induced bodyweight gain in Chinese Han population. Prog Neuro-Psychoph. 2013;43:197–202.
Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–48.
Zhang W, Rohlmann A, Sargsyan V, Aramuni G, Hammer RE, Sudhof TC, et al. Extracellular domains of alpha-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. J Neurosci. 2005;25:4330–42.
Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–8003.
Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502.
Mena A, Ruiz-Salas JC, Puentes A, Dorado I, De Ruiz-Veguilla M, et al. Reduced prepulse inhibition as a biomarker of schizophrenia. Front Behav Neurosci. 2016;10:202.
Grayton HM, Missler M, Collier DA, Fernandes C. Altered social behaviours in neurexin 1alpha knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One. 2013;8:e67114.
Sons MS, Busche N, Strenzke N, Moser T, Ernsberger U, Mooren FC, et al. alpha-Neurexins are required for efficient transmitter release and synaptic homeostasis at the mouse neuromuscular junction. Neurosci. 2006;138:433–46.
Dudanova I, Tabuchi K, Rohlmann A, Sudhof TC, Missler M. Deletion of alpha-neurexins does not cause a major impairment of axonal pathfinding or synapse formation. J Comp Neurol. 2007;502:261–74.
Dachtler J, Glasper J, Cohen RN, Ivorra JL, Swiffen DJ, Jackson AJ, et al. Deletion of alpha-neurexin II results in autism-related behaviors in mice. Transl Psychiatry. 2014;4:e484.
Dachtler J, Ivorra JL, Rowland TE, Lever C, Rodgers RJ, Clapcote SJ. Heterozygous deletion of alpha-neurexin I or alpha-neurexin II results in behaviors relevant to autism and schizophrenia. Behav Neurosci. 2015;129:765–76.
Born G, Grayton HM, Langhorst H, Dudanova I, Rohlmann A, Woodward BW, et al. Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front Synaptic Neurosci. 2015;7:3.
Rabaneda LG, Robles-Lanuza E, Nieto-Gonzalez JL, Scholl FG. Neurexin dysfunction in adult neurons results in autistic-like behavior in mice. Cell Rep. 2014;8:338–46.
Anderson GR, Aoto J, Tabuchi K, Foldy C, Covy J, Yee AX, et al. beta-neurexins control neural circuits by regulating Synaptic endocannabinoid signaling. Cell. 2015;162:593–606.
Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–61.
Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–79.
Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–45.
Boucard AA, Ko J, Sudhof TC. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem. 2012;287:9399–413.
Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–8.
Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci. 2010;30:7495–506.
Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Sudhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88.
Dai J, Aoto J, Sudhof TC. Alternative splicing of presynaptic neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron. 2019;102:993–1008 e1005.
Aoto J, Foldy C, Ilcus SM, Tabuchi K, Sudhof TC. Distinct circuit-dependent functions of presynaptic neurexin-3 at GABAergic and glutamatergic synapses. Nat Neurosci. 2015;18:997–1007.
Chen LY, Jiang M, Zhang B, Gokce O, Sudhof TC. Conditional deletion of all neurexins defines diversity of essential synaptic organizer functions for neurexins. Neuron 2017;94:611–25 e614.
Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:693–7.
De Los Angeles A, Tunbridge EM. Unraveling mechanisms of patient-specific NRXN1 mutations in neuropsychiatric diseases using human induced pluripotent stem cells. Stem Cells Dev. 2020;29:1142–4.
Flaherty E, Zhu S, Barretto N, Cheng E, Deans PJM, Fernando MB, et al. Neuronal impact of patient-specific aberrant NRXN1alpha splicing. Nat Genet. 2019;51:1679–90.
Fuccillo MV, Foldy C, Gokce O, Rothwell PE, Sun GL, Malenka RC, et al. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–40.
Jahn K, Heese A, Kebir O, Groh A, Bleich S, Krebs MO, et al. Differential methylation pattern of schizophrenia candidate genes in tetrahydrocannabinol-consuming treatment-resistant schizophrenic patients compared to non-consumer patients and healthy controls. Neuropsychobiology. 2020;Jun 29:1–9.
Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, Gelernter J, et al. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17:1569–77.
Hishimoto A, Liu QR, Drgon T, Pletnikova O, Walther D, Zhu XG, et al. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum Mol Genet. 2007;16:2880–91.
Panagopoulos VN, Trull TJ, Glowinski AL, Lynskey MT, Heath AC, Agrawal A, et al. Examining the association of NRXN3 SNPs with borderline personality disorder phenotypes in heroin dependent cases and socio-economically disadvantaged controls. Drug Alcohol Depend. 2013;128:187–93.
Stoltenberg SF, Lehmann MK, Christ CC, Hersrud SL, Davies GE. Associations among types of impulsivity, substance use problems and neurexin-3 polymorphisms. Drug Alcohol Depend. 2011;119:e31–38.
Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–9.
Sarachana T, Hu VW. Genome-wide identification of transcriptional targets of RORA reveals direct regulation of multiple genes associated with autism spectrum disorder. Mol Autism. 2013;4:14.
El-Brolosy MA, Stainier DYR. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 2017;13:e1006780.
El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568:193–7.
Ma ZP, Chen J. Nonsense mutations and genetic compensation response. Yi Chuan. 2019;41:359–64.
Sztal TE, Stainier DYR. Transcriptional adaptation: a mechanism underlying genetic robustness. Development. 2020;147.
Trotter JH, Hao J, Maxeiner S, Tsetsenis T, Liu Z, Zhuang X, et al. Synaptic neurexin-1 assembles into dynamically regulated active zone nanoclusters. J Cell Biol. 2019;218:2677–98.
Avazzadeh S, McDonagh K, Reilly J, Wang Y, Boomkamp SD, McInerney V, et al. Increased Ca(2+) signaling in NRXN1alpha (+/−) neurons derived from ASD induced pluripotent stem cells. Mol Autism. 2019;10:52.
Lam M, Moslem M, Bryois J, Pronk RJ, Uhlin E, Ellstrom ID, et al. Single cell analysis of autism patient with bi-allelic NRXN1-alpha deletion reveals skewed fate choice in neural progenitors and impaired neuronal functionality. Exp Cell Res. 2019;383:111469.
Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S, et al. Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in NRXN1. Cell Stem Cell. 2015;17:316–28.
Zeng L, Zhang P, Shi L, Yamamoto V, Lu W, Wang K. Functional impacts of NRXN1 knockdown on neurodevelopment in stem cell models. PLoS One. 2013;8:e59685.
Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36.
Giacomotto J, Rinkwitz S, Becker TS. Effective heritable gene knockdown in zebrafish using synthetic microRNAs. Nat Commun. 2015;6:7378.
Giacomotto J, Carroll AP, Rinkwitz S, Mowry B, Cairns MJ, Becker TS. Developmental suppression of schizophrenia-associated miR-137 alters sensorimotor function in zebrafish. Transl Psychiatry. 2016;6:e818.
Periyasamy S, John S, Padmavati R, Rajendren P, Thirunavukkarasu P, Gratten J et al. Association of schizophrenia risk with disordered niacin metabolism in an indian genome-wide association study. JAMA Psychiatry. 2019;76:1026–34.
Laird AS, Mackovski N, Rinkwitz S, Becker TS, Giacomotto J. Tissue-specific models of spinal muscular atrophy confirm a critical role of SMN in motor neurons from embryonic to adult stages. Hum Mol Genet. 2016;25:1728–38.
Forster D, Kramer A, Baier H, Kubo F. Optogenetic precision toolkit to reveal form, function and connectivity of single neurons. Methods 2018;150:42–48.
Forster D, Dal Maschio M, Laurell E, Baier H. An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits. Nat Commun. 2017;8:116.
Ji N, Freeman J, Smith SL. Technologies for imaging neural activity in large volumes. Nat Neurosci. 2016;19:1154–64.
Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10:413.
Al Shehhi M, Forman EB, Fitzgerald JE, McInerney V, Krawczyk J, Shen S, et al. NRXN1 deletion syndrome; phenotypic and penetrance data from 34 families. Eur J Med Genet. 2019;62:204–9.
Rochtus AM, Trowbridge S, Goldstein RD, Sheidley BR, Prabhu SP, Haynes R, et al. Mutations in NRXN1 and NRXN2 in a patient with early-onset epileptic encephalopathy and respiratory depression. Cold Spring Harb Mol Case Stud. 2019;5:1–13.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006.
Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–8.
Acknowledgements
This work was supported by grants from the Australian National Health and Medical Research Council (NHMRC) Project Grant no. 1165850 to BM and JG, The Rebecca Cooper Medical Research Project Grant no. PG2019405 to JG. JG was also supported by a NHMRC Emerging Leader Fellowship no. 1174145.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tromp, A., Mowry, B. & Giacomotto, J. Neurexins in autism and schizophrenia—a review of patient mutations, mouse models and potential future directions. Mol Psychiatry 26, 747–760 (2021). https://doi.org/10.1038/s41380-020-00944-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-00944-8
This article is cited by
-
Morphological and transcriptomic analyses of stem cell-derived cortical neurons reveal mechanisms underlying synaptic dysfunction in schizophrenia
Genome Medicine (2023)
-
NRXN1 depletion in the medial prefrontal cortex induces anxiety-like behaviors and abnormal social phenotypes along with impaired neurite outgrowth in rat
Journal of Neurodevelopmental Disorders (2023)
-
Effect of schizophrenia common variants on infant brain volumes: cross-sectional study in 207 term neonates in developing Human Connectome Project
Translational Psychiatry (2023)
-
Lineage-specific regulatory changes in hypertrophic cardiomyopathy unraveled by single-nucleus RNA-seq and spatial transcriptomics
Cell Discovery (2023)
-
Dysregulation of Synaptic Plasticity Markers in Schizophrenia
Indian Journal of Clinical Biochemistry (2023)