Abstract
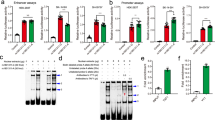
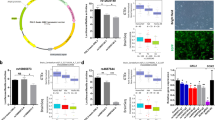
Schizophrenia (SCZ) is a neuropsychiatric disorder with aberrant expression of multiple genes. However, identifying its exact causal genes remains a considerable challenge. The brain-specific transcription factor POU3F2 (POU domain, class 3, transcription factor 2) has been recognized as a risk factor for SCZ, but our understanding of its target genes and pathogenic mechanisms are still limited. Here we report that POU3F2 regulates 42 SCZ-related genes in knockdown and RNA-sequencing experiments of human neural progenitor cells (NPCs). Among those SCZ-related genes, TRIM8 (Tripartite motif containing 8) is located in SCZ-associated genetic locus and is aberrantly expressed in patients with SCZ. Luciferase reporter and electrophoretic mobility shift assays (EMSA) showed that POU3F2 induces TRIM8 expression by binding to the SCZ-associated SNP (single nucleotide polymorphism) rs5011218, which affects POU3F2-binding efficiency at the promoter region of TRIM8. We investigated the cellular functions of POU3F2 and TRIM8 as they co-regulate several pathways related to neural development and synaptic function. Knocking down either POU3F2 or TRIM8 promoted the proliferation of NPCs, inhibited their neuronal differentiation, and impaired the excitatory synaptic transmission of NPC-derived neurons. These results indicate that POU3F2 regulates TRIM8 expression through the SCZ-associated SNP rs5011218, and both genes may be involved in the etiology of SCZ by regulating neural development and synaptic function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Walker E, Kestler L, Bollini A, Hochman KM. Schizophrenia: etiology and course. Annu Rev Psychol. 2004;55:401–30.
Frankle WG, Lerma J, Laruelle M. The synaptic hypothesis of schizophrenia. Neuron. 2003;39:205–16.
Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017;18:727–40.
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Yang CP, Li X, Wu Y, Shen Q, Zeng Y, Xiong Q, et al. Comprehensive integrative analyses identify GLT8D1 and CSNK2B as schizophrenia risk genes. Nat Commun. 2018;9:838.
Yin J, Lin J, Luo X, Chen Y, Li Z, Ma G, et al. miR-137: a new player in schizophrenia. Int J Mol Sci. 2014;15:3262–71.
Simms BA, Zamponi GW. Neuronal voltage-gated calcium channels: structure, function, and dysfunction. Neuron. 2014;82:24–45.
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Chen C, Meng Q, Xia Y, Ding C, Wang L, Dai R, et al. The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci Transl Med. 2018;10:eaat8178.
Ren Y, Cui Y, Li X, Wang B, Na L, Shi J, et al. A co-expression network analysis reveals lncRNA abnormalities in peripheral blood in early-onset schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2015;63:1–5.
He X, Treacy M, Simmons D, Ingraham H, Swanson L, Rosenfeld M. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989;340:35–41.
McEvilly R, de DM, Schonemann M, Hooshmand F, Rosenfeld M. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–32.
Hashizume K, Yamanaka M, Ueda S. POU3F2 participates in cognitive function and adult hippocampal neurogenesis via mammalian-characteristic amino acid repeats. Genes Brain Behav. 2018;17:118–25.
Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, et al. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 2009;35:96–108.
Pearl JR, Colantuoni C, Bergey DE, Funk CC, Shannon P, Basu B, et al. Genome-scale transcriptional regulatory network models of psychiatric and neurodegenerative disorders. Cell Syst. 2019;8:122–35. e127.
Li Q, Yan J, Mao AP, Li C, Ran Y, Shu HB, et al. Tripartite motif 8 (TRIM8) modulates TNFalpha- and IL-1beta-triggered NF-kappaB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci USA. 2011;108:19341–6.
Caratozzolo MF, Micale L, Turturo MG, Cornacchia S, Fusco C, Marzano F, et al. TRIM8 modulates p53 activity to dictate cell cycle arrest. Cell Cycle. 2012;11:511–23.
Wu Y, Yao YG, Luo XJ. SZDB: a database for schizophrenia genetic research. Schizophr Bull. 2017;43:459–71.
Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127.
Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, et al. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–29.
Huo Y, Li S, Liu J, Li X, Luo XJ. Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nat Commun. 2019;10:670.
Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–87.
Glass D, Vinuela A, Davies MN, Ramasamy A, Parts L, Knowles D, et al. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 2013;14:R75.
Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21:1117–25.
Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63.
Xue Y, Qian H, Hu J, Zhou B, Zhou Y, Hu X, et al. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat Neurosci. 2016;19:807–15.
Consortium GT. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–5.
Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci USA. 2003;100:6347–52.
Cui L, Sun W, Yu M, Li N, Guo L, Gu H, et al. Disrupted-in-schizophrenia1 (DISC1) L100P mutation alters synaptic transmission and plasticity in the hippocampus and causes recognition memory deficits. Mol Brain. 2016;9:89.
Mao W, Watanabe T, Cho S, Frost JL, Truong T, Zhao X, et al. Shank1 regulates excitatory synaptic transmission in mouse hippocampal parvalbumin-expressing inhibitory interneurons. Eur J Neurosci. 2015;41:1025–35.
Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Whitehead Pavlides JM, et al. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018;173:1705–15.
Rosso SB, Inestrosa NC. WNT signaling in neuronal maturation and synaptogenesis. Front Cell Neurosci. 2013;7:103.
Isabella P, Flavia N, Alberto MF, Giorgio DK, Antonio DC, Chiara R, et al. Neurodevelopment in schizophrenia: The role of the Wnt pathways. Curr Neuropharmacol. 2013;11:535–58.
Massey PV, Bashir ZI. Long-term depression: multiple forms and implications for brain function. Trends Neurosci. 2007;30:176–84.
Hasan A, Nitsche MA, Herrmann M, Schneider-Axmann T, Marshall L, Gruber O, et al. Impaired long-term depression in schizophrenia: a cathodal tDCS pilot study. Brain Stimul. 2012;5:475–83.
Russell SA, Bashaw GJ. Axon guidance pathways and the control of gene expression. Dev Dyn. 2018;247:571–80.
Klein R, Kania A. Ephrin signalling in the developing nervous system. Curr Opin Neurobiol. 2014;27:16–24.
Liu YN, Lu SY, Yao J. Application of induced pluripotent stem cells to understand neurobiological basis of bipolar disorder and schizophrenia. Psychiatry Clin Neurosci. 2017;71:579–99.
Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–5.
Yoon K-J, Nguyen HaN, Ursini G, Zhang F, Kim N-S, Wen Z, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91.
Habela CW, Song H, Ming GL. Modeling synaptogenesis in schizophrenia and autism using human iPSC derived neurons. Mol Cell Neurosci. 2016;73:52–62.
Wang L, Lockstone HE, Guest PC, Levin Y, Palotás A, Pietsch S, et al. Expression profiling of fibroblasts identifies cell cycle abnormalities in schizophrenia. J Proteome Res. 2010;9:521–7.
Allen KM, Fung SJ, Weickert CS. Cell proliferation is reduced in the hippocampus in schizophrenia. Aust N Z J Psychiatry. 2016;50:473–80.
Sanches M, Keshavan MS, Brambilla P, Soares JC. Neurodevelopmental basis of bipolar disorder: a critical appraisal. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1617–27.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Stephan KE, Friston KJ, Frith CD. Dysconnection in schizophrenia: from abnormal synaptic plasticity to failures of self-monitoring. Schizophr Bull. 2009;35:509–27.
Wong AHC, Van Tol HHM. Schizophrenia: from phenomenology to neurobiology. Neurosci Biobehav Rev. 2003;27:269–306.
Karlsgodt KH, Sun D, Cannon TD. Structural and functional brain abnormalities in schizophrenia. Curr Dir Psychol Sci. 2010;19:226–31.
Westphal DS, Riedhammer KM, Kovacs-Nagy R, Meitinger T, Hoefele J, Wagner M. A de novo missense variant in POU3F2 identified in a child with global developmental delay. Neuropediatrics. 2018;49:401–4.
Kasher PR, Schertz KE, Thomas M, Jackson A, Annunziata S, Ballesta-Martinez MJ, et al. Small 6q16.1 deletions encompassing POU3F2 cause susceptibility to obesity and variable developmental delay with intellectual disability. Am J Hum Genet. 2016;98:363–72.
Assoum M, Lines MA, Elpeleg O, Darmency V, Whiting S, Edvardson S, et al. Further delineation of the clinical spectrum of de novo TRIM8 truncating mutations. Am J Med Genet A. 2018;176:2470–8.
Sakai Y, Fukai R, Matsushita Y, Miyake N, Saitsu H, Akamine S, et al. De novo truncating mutation of TRIM8 causes early-onset epileptic encephalopathy. Ann Hum Genet. 2016;80:235–40.
Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23:2632–43.
Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–44.
Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108:10343–8.
Venuto S, Castellana S, Monti M, Appolloni I, Fusilli C, Fusco C, et al. TRIM8-driven transcriptomic profile of neural stem cells identified glioma-related nodal genes and pathways. Biochim Biophys Acta Gen Subj. 2019;1863:491–501.
Kohlbrenner EA, Shaskan N, Pietersen CY, Sonntag KC, Woo TW. Gene expression profile associated with postnatal development of pyramidal neurons in the human prefrontal cortex implicates ubiquitin ligase E3 in the pathophysiology of schizophrenia onset. J Psychiatr Res. 2018;102:110–7.
Ma Q, Ruan H, Peng L, Zhang M, Gack MU, Yao WD. Proteasome-independent polyubiquitin linkage regulates synapse scaffolding, efficacy, and plasticity. Proc Natl Acad Sci USA. 2017;114:E8760–9.
van Kesteren CF, Gremmels H, de Witte LD, Hol EM, Van Gool AR, Falkai PG, et al. Immune involvement in the pathogenesis of schizophrenia: a meta-analysis on postmortem brain studies. Transl Psychiatry. 2017;7:e1075.
Georgala PA, Carr CB, Price DJ. The role of Pax6 in forebrain development. Dev Neurobiol. 2011;71:690–709.
Zamani A, Xiao J, Turnley AM, Murray SS. Tropomyosin-related kinase B (TrkB) regulates neurite outgrowth via a novel interaction with suppressor of cytokine signalling 2 (SOCS2). Mol Neurobiol. 2019;56:1262–75.
Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–42.
Takahashi K, Yamada M, Ohata H, Honda K, Yamada M. Ndrg2 promotes neurite outgrowth of NGF-differentiated PC12 cells. Neurosci Lett. 2005;388:157–62.
Lin K, Yin A, Yao L, Li Y. N-myc downstream-regulated gene 2 in the nervous system: from expression pattern to function. Acta Biochim Biophys Sin. 2015;47:761–6.
Christa KP, Len AP, Kimberly L, Wufang F, Gregory GL. Characterization of the human neurocan gene, CSPG3. Gene. 1998;221:199–205.
Logan S, Arzua T, Canfield SG, Seminary ER, Sison SL, Ebert AD, et al. Studying human neurological disorders using induced pluripotent stem cells: from 2D monolayer to 3D organoid and blood brain barrier models. Compr Physiol. 2019;9:565–611.
Hu Z, Zhou M, Wu Y, Li Z, Liu X, Wu L, et al. ssODN-mediated in-frame deletion with CRISPR/Cas9 to restore the FVIII function in hemophilia A patient-derived iPSCs and ECs. Mol Ther Nucleic Acids. 2019;17:198–209.
Andrews S. FastQC: a quality control tool for high throughput sequence data. Cambridge: Babraham Bioinformatics, Babraham Institute; 2010.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–30.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Popko J, Fernandes A, Brites D, Lanier LM. Automated analysis of NeuronJ tracing data. Cytom A. 2009;75:371–6.
Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137.
Acknowledgements
We thank Glatt’s lab at Upstate Medical University for providing technical support.
Funding
This work was supported by National Natural Science Foundation of China (NSFC) grants 31970572, 31871276, 31571312, and 81401114, National Key R&D Project of China 2016YFC1306000 and 2017YFC0908701, Innovation-driven Project of Central South University 2020CX003 (to CC) and NIH grants 1U01MH103340, 1U01MH116489, and 1R01MH110920, New York State Empire Innovation Program (to CL).
Author information
Authors and Affiliations
Contributions
CL and CC designed and guided the study. CD, LW, and QM performed the gene knockdown, NPC proliferation, dual-luciferase reporter and neuronal differentiation assays. CZ, YX, YJ, SM, and RD analyzed the RNA-seq data and ran the enrichment analysis. HR did the electrophysiological recordings. CD wrote the manuscript with substantive edits from RK, LK, MW, WY, CL, and CC.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ding, C., Zhang, C., Kopp, R. et al. Transcription factor POU3F2 regulates TRIM8 expression contributing to cellular functions implicated in schizophrenia. Mol Psychiatry 26, 3444–3460 (2021). https://doi.org/10.1038/s41380-020-00877-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-00877-2
This article is cited by
-
Gatad2b, associated with the neurodevelopmental syndrome GAND, plays a critical role in neurodevelopment and cortical patterning
Translational Psychiatry (2024)
-
Transition of allele-specific DNA hydroxymethylation at regulatory loci is associated with phenotypic variation in monozygotic twins discordant for psychiatric disorders
BMC Medicine (2023)
-
A higher dysregulation burden of brain DNA methylation in female patients implicated in the sex bias of Schizophrenia
Molecular Psychiatry (2023)
-
The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases
BMC Medical Genomics (2022)
-
Phenotypes, mechanisms and therapeutics: insights from bipolar disorder GWAS findings
Molecular Psychiatry (2022)