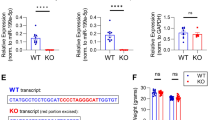
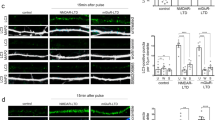
Abstract
Synaptic plasticity requires a tight control of mRNA levels in dendrites. RNA translation and degradation pathways have been recently linked to neurodevelopmental and neuropsychiatric diseases, suggesting a role for RNA regulation in synaptic plasticity and cognition. While the local translation of specific mRNAs has been implicated in synaptic plasticity, the tightly controlled mechanisms that regulate local quantity of specific mRNAs remain poorly understood. Despite being the only RNA regulatory pathway that is associated with multiple mental illnesses, the nonsense-mediated mRNA decay (NMD) pathway presents an unexplored regulatory mechanism for synaptic function and plasticity. Here, we show that neuron-specific disruption of UPF2, an NMD component, in adulthood attenuates learning, memory, spine density, synaptic plasticity (L-LTP), and potentiates perseverative/repetitive behavior in mice. We report that the NMD pathway operates within dendrites to regulate Glutamate Receptor 1 (GLUR1) surface levels. Specifically, UPF2 modulates the internalization of GLUR1 and promotes its local synthesis in dendrites. We identified neuronal Prkag3 mRNA as a mechanistic substrate for NMD that contributes to the UPF2-mediated regulation of GLUR1 by limiting total GLUR1 levels. These data establish that UPF2 regulates synaptic plasticity, cognition, and local protein synthesis in dendrites, providing fundamental insight into the neuron-specific function of NMD within the brain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–8.
Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode caenorhabditis elegans. Genetics. 1989;123:301–13.
Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–77.
Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–15.
Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–41.
Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27 3 Pt 2:543–53.
Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–9003.
Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–83.
Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim Biophys Acta. 2013;1829:624–33.
Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–8.
Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–65.
Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21:394–402.
Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–91.
Fujimoto T, Tanaka H, Kumamaru E, Okamura K, Miki N. Arc interacts with microtubules/microtubule-associated protein 2 and attenuates microtubule-associated protein 2 immunoreactivity in the dendrites. J Neurosci Res. 2004;76:51–63.
Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–9.
Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML. Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience. 2004;125:7–11.
Rodriguez JJ, Davies HA, Silva AT, De Souza IE, Peddie CJ, Colyer FM, et al. Long-term potentiation in the rat dentate gyrus is associated with enhanced Arc/Arg3.1 protein expression in spines, dendrites and glia. Eur J Neurosci. 2005;21:2384–96.
Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, et al. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001.
Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–44.
Smith-Hicks C, Xiao B, Deng R, Ji Y, Zhao X, Shepherd JD, et al. SRF binding to SRE 6.9 in the Arc promoter is essential for LTD in cultured Purkinje cells. Nat Neurosci. 2010;13:1082–9.
Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97.
Cajigas IJ, Tushev G, Will TJ, Dieck Stom, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–66.
Brown TC, Tran IC, Backos DS, Esteban JA. NMDA receptor-dependent activation of the small GTPase Rab5 drives the removal of synaptic AMPA receptors during hippocampal LTD. Neuron. 2005;45:81–94.
Fu AK, Hung KW, Fu WY, Shen C, Chen Y, Xia J, et al. APC(Cdh1) mediates EphA4-dependent downregulation of AMPA receptors in homeostatic plasticity. Nat Neurosci. 2011;14:181–9.
Han Y, Luo Y, Sun J, Ding Z, Liu J, Yan W, et al. AMPK signaling in the dorsal hippocampus negatively regulates contextual fear memory formation. Neuropsychopharmacology. 2016;41:1849–64.
Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14:279–84.
Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, et al. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2011;16:238–9.
Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry. 2010;15:767–76.
Lynch SA, Nguyen LS, Ng LY, Waldron M, McDonald D, Gecz J. Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur J Med Genet. 2012;55:476–9.
Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–33.
Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22:1816–25.
Xu X, Zhang L, Tong P, Xun G, Su W, Xiong Z, et al. Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin Genet. 2013;83:560–4.
Long AA, Mahapatra CT, Woodruff EA, Rohrbough J, Leung H-T, Shino S, et al. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci. 2010;123:3303–15.
Mooney CM, Jimenez-Mateos EM, Engel T, Mooney C, Diviney M, Venø MT, et al. RNA sequencing of synaptic and cytoplasmic Upf1-bound transcripts supports contribution of nonsense-mediated decay to epileptogenesis. Sci Rep. 2017;7:41517.
Huang L, Shum E, Jones S, Lou C, Dumdie J, Kim H, et al. A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol Psychiatry. 2018;23:1773.
Karam R, Wilkinson M. A conserved microRNA/NMD regulatory circuit controls gene expression. RNA Biol. 2012;9:22–6.
Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21.
Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci. 2012;35:417–43.
Crabtree GW, Gogos JA. Synaptic plasticity, neural circuits, and the emerging role of altered short-term information processing in schizophrenia. Front Synaptic Neurosci. 2014;6:28.
Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, et al. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–55.
Bassell GJ, Kelic S. Binding proteins for mRNA localization and local translation, and their dysfunction in genetic neurological disease. Curr Opin Neurobiol. 2004;14:574–81.
Buffington SA, Huang W, Costa-Mattioli M. Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci. 2014;37:17–38.
Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–6.
Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–7.
Kelleher RJ 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–6.
Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, et al. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–93.
Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–5.
Huber KM, Klann E, Costa-Mattioli M, Zukin RS. Dysregulation of mammalian target of rapamycin signaling in mouse models of autism. J Neurosci. 2015;35:13836–42.
Sidorov MS, Auerbach BD, Bear MF. Fragile X mental retardation protein and synaptic plasticity. Mol Brain. 2013;6:15.
Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–96.
Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–8.
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40.
Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–50.
Korb E, Finkbeiner S. Arc in synaptic plasticity: from gene to behavior. Trends Neurosci. 2011;34:591–8.
Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–93.
Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–43.
Le Hir H, Sauliere J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol. 2016;17:41–54.
Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrançois M, Chagnon P, et al. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143:251–62.
Fleming JJ, England PM. AMPA receptors and synaptic plasticity: a chemist’s perspective. Nat Chem Biol. 2010;6:89–97.
Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol. 2012;22:461–9.
He F, Jacobson A. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu Rev Genet. 2015;49:339–66.
Korb E, Wilkinson CL, Delgado RN, Lovero KL, Finkbeiner S. Arc in the nucleus regulates PML-dependent GluA1 transcription and homeostatic plasticity. Nat Neurosci. 2013;16:874.
Soulé J, Alme M, Myrum C, Schubert M, Kanhema T, Bramham CR. Balancing arc synthesis, mRNA decay, and proteasomal degradation maximal protein expression triggered by rapid eye movement sleep-like bursts of muscarinic cholinergic receptor stimulation. J Biol Chem. 2012;287:22354–66.
Farris S, Lewandowski G, Cox CD, Steward O. Selective localization of arc mRNA in dendrites involves activity-and translation-dependent mRNA degradation. J Neurosci. 2014;34:4481–93.
Ninomiya K, Ohno M, Kataoka N. Dendritic transport element of human arc mRNA confers RNA degradation activity in a translation‐dependent manner. Genes Cells. 2016;21:1263–9.
Paolantoni C, Ricciardi S, De Paolis V, Okenwa C, Catalanotto C, Ciotti MT, et al. Arc 3’UTR splicing leads to dual and antagonistic effects in fine-tuning Arc expression upon BDNF signaling. Front Mol Neurosci. 2018;11:145.
Steward O, Matsudaira Yee K, Farris S, Pirbhoy PS, Worley P, Okamura K, et al. Delayed degradation and impaired dendritic delivery of intron-lacking EGFP-Arc/Arg3. 1 mRNA in EGFP-Arc transgenic mice. Front Mol Neurosci. 2018;10:435.
Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–59.
Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Sudhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88.
Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–74.
Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, et al. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–53.
Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F, et al. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J Neurochem. 2011;119:27–39.
Schwarz LA, Hall BJ, Patrick GN. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J Neurosci. 2010;30:16718–29.
Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605.
Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–25.
Patrick GN, Bingol B, Weld HA, Schuman EM. Ubiquitin-mediated proteasome activity is required for agonist-induced endocytosis of GluRs. Curr Biol. 2003;13:2073–81.
Widagdo J, YJ Chai, MC Ridder, YQ Chau, RC Johnson, P Sah, et al. Activity-dependent ubiquitination of GluA1 and GluA2 regulates AMPA receptor intracellular sorting and degradation. Cell Rep. 2015;10:783–95.
Zhang D, Hou Q, Wang M, Lin A, Jarzylo L, Navis A, et al. Na,K-ATPase activity regulates AMPA receptor turnover through proteasome-mediated proteolysis. J Neurosci. 2009;29:4498–511.
Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci USA. 2006;103:9482–7.
Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–17.
Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J. Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol Cell. 2007;27:780–92.
Chamieh H, Ballut L, Bonneau F, Hir HLe. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93.
Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–61.
Deglincerti A, Jaffrey SR. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:120079.
Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98.
Taylor AM, Berchtold NC, Perreau VM, Tu CH, Jeon NLi, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–707.
Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22:4673–87.
Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, et al. An alternative branch of the nonsense‐mediated decay pathway. EMBO J. 2007;26:1820–30.
Karam R, CH Lou, H Kroeger, L Huang, JH Lin, MF Wilkinson, The unfolded protein response is shaped by the NMD pathway. EMBO Rep; 2015;16:599–609.
Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26.
Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277:23977–80.
Inoki K, Zhu T, Guan K-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90.
Horman S, Browne GJ, Krause U, Patel JV, Vertommen D, Bertrand L, et al. Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Curr Biol. 2002;12:1419–23.
Kimura N, Tokunaga C, Dalal S, Richardson C, Yoshino Ki, Hara K, et al. A possible linkage between AMP‐activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells. 2003;8:65–79.
Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603.
Ma T, Chen Y, Vingtdeux V, Zhao H, Viollet B, Marambaud P, et al. Inhibition of AMP-activated protein kinase signaling alleviates impairments in hippocampal synaptic plasticity induced by amyloid β. J Neurosci. 2014;34:12230–8.
Matsuo N, Reijmers L, Mayford M. Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science. 2008;319:1104–7.
Whitlock JR, Heynen AJ, Shuler MG, Bear MF. Learning induces long-term potentiation in the hippocampus. Science. 2006;313:1093–7.
Reisel D, Bannerman DM, Schmitt WB, Deacon RM, Flint J, Borchardt T, et al. Spatial memory dissociations in mice lacking GluR1. Nat Neurosci. 2002;5:868–73.
Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–27.
Swanger SA, He YA, Richter JD, Bassell GJ. Dendritic GluN2A synthesis mediates activity-induced NMDA receptor insertion. J Neurosci. 2013;33:8898–908.
Farris S, Lewandowski G, Cox CD, Steward O. Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J Neurosci. 2014;34:4481–93.
Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–7.
Gu Z, Jiang Q, Fu AK, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25:4974–84.
Noel J, Ralph GS, Pickard L, Williams J, Molnar E, Uney JB, et al. Surface expression of AMPA receptors in hippocampal neurons is regulated by an NSF-dependent mechanism. Neuron. 1999;23:365–76.
Lin JW, Ju W, Foster K, Lee SH, Ahmadian G, Wyszynski M, et al. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat Neurosci. 2000;3:1282–90.
Ning X, Guo J, Wolfert MA, Boons GJ. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew Chem Int Ed Engl. 2008;47:2253–5.
Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–7.
Notaras M, Du X, Gogos J, Van Den Buuse M, Hill R. The BDNF Val66Met polymorphism regulates glucocorticoid-induced corticohippocampal remodeling and behavioral despair. Transl Psychiatry. 2017;7:e1233.
Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–3.
Trinh MA, Ma T, Kaphzan H, Bhattacharya A, Antion MD, Cavener DR, et al. The eIF2a kinase PERK limits the expression of hippocampal metabotropic glutamate receptor-dependent long-term depression. Learn Mem. 2014;21:298–304.
Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, et al. Dysregulation of mTOR signaling in fragile x syndrome. J Neurosci. 2010;30:694–702.
Sarnyai Z, Sibille EL, Pavlides C, Fenster RJ, McEwen BS, Toth M. Impaired hippocampal-dependent learning and functional abnormalities in the hippocampus in mice lacking serotonin(1A) receptors. Proc Natl Acad Sci USA. 2000;97:14731–6.
Notaras M, Hill R, Gogos J, van den Buuse M. BDNF Val66Met genotype determines hippocampus-dependent behavior via sensitivity to glucocorticoid signaling. Mol Psychiatry. 2016;21:730–2.
Dumont M, Wille E, Stack C, Calingasan NY, Beal MF, Lin MT. Reduction of oxidative stress, amyloid deposition, and memory deficit by manganese superoxide dismutase overexpression in a transgenic mouse model of Alzheimer’s disease. FASEB J. 2009;23:2459–66.
Angoa-Perez M, Kane MJ, Briggs DI, Francescutti DM, Kuhn DM. Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice. J Vis Exp. 2013;82:50978.
Silverman JL, Tolu SS, Barkan CL, Crawley JN. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2010;35:976–89.
Mitchell E, Klein SL, Argyropoulos KV, Sharma A, Chan RB, Toth JG, et al. Behavioural traits propagate across generations via segregated iterative-somatic and gametic epigentic mechanisms. Nat Commun. 2016;7:11492.
Walf AA, Frye CA. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat Protoc. 2007;2:322–8.
Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302.
Notaras M, Hill R, Gogos J, van den Buuse M. BDNF Val66Met genotype interacts with a history of simulated stress exposure to regulate sensorimotor gating and startle reactivity. Schizophr Bull. 2017;43:665–72.
Acknowledgements
We thank J. Lykke-Andersen for generously providing the anti-UPF1 and anti-UPF2 antibodies, M.E. Ross for helpful comments and suggestions and M. Toth for helpful suggestions regarding behavioral assays. This work was supported by a NHMRC CJ Martin Biomedical Fellowship awarded to MN, KoreaNRF-2015R1A2A1A09005662 to NLJ, NIH grants NS034007 and NS047384 to EK, and NIH R01 MH114888 and Leon Levy Foundation Grants to DC.
Author information
Authors and Affiliations
Contributions
MN designed, performed, and/or analyzed most of the experiments including in vivo assays, as well as manuscript preparation/writing. MA prepared all neuronal cultures. FL performed electrophysiology experiments. NV provided technical assistance. MT provided infrastructure support. NLJ designed and manufactured microfluidic devices. EK supervised electrophysiology experiments. DC initiated and conceived the project; designed, analyzed, and supervised experiments; and wrote the manuscript. All authors contributed to the experimental design and interpretation and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Notaras, M., Allen, M., Longo, F. et al. UPF2 leads to degradation of dendritically targeted mRNAs to regulate synaptic plasticity and cognitive function. Mol Psychiatry 25, 3360–3379 (2020). https://doi.org/10.1038/s41380-019-0547-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0547-5
This article is cited by
-
FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells
Genome Biology (2024)
-
Analysis of subcellular RNA fractions demonstrates significant genetic regulation of gene expression in human brain post-transcriptionally
Scientific Reports (2023)
-
Circular RNA: A novel and potential regulator in pathophysiology of schizophrenia
Metabolic Brain Disease (2022)
-
NMD abnormalities during brain development in the Fmr1-knockout mouse model of fragile X syndrome
Genome Biology (2021)
-
Neurodevelopmental signatures of narcotic and neuropsychiatric risk factors in 3D human-derived forebrain organoids
Molecular Psychiatry (2021)