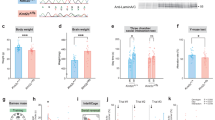
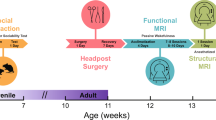
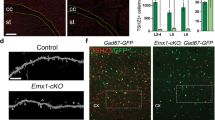
Abstract
Cullin 3 (Cul3) gene, which encodes a core component of the E3 ubiquitin ligase complex that mediates proteasomal degradation, has been identified as a true high-risk factor for autism. Here, by combining behavioral, electrophysiological, and proteomic approaches, we have examined how Cul3 deficiency contributes to the etiology of different aspects of autism. Heterozygous mice with forebrain Cul3 deletion displayed autism-like social interaction impairment and sensory-gating deficiency. Region-specific deletion of Cul3 leads to distinct phenotypes, with social deficits linked to the loss of Cul3 in prefrontal cortex (PFC), and stereotypic behaviors linked to the loss of Cul3 in striatum. Correlated with these behavioral alterations, Cul3 deficiency in forebrain or PFC induces NMDA receptor hypofunction, while Cul3 loss in striatum causes a cell type-specific alteration of neuronal excitability in striatal circuits. Large-scale profiling has identified sets of misregulated proteins resulting from Cul3 deficiency in different regions, including Smyd3, a histone methyltransferase involved in gene transcription. Inhibition or knockdown of Smyd3 in forebrain Cul3-deficient mice ameliorates social deficits and restores NMDAR function in PFC. These results have revealed for the first time a potential molecular mechanism underlying the manifestation of different autism-like behavioral deficits by Cul3 deletion in cortico-striatal circuits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Proteomic data will be deposited in a public repository. The access number and the dataset will be available for access.
References
de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22:345–61.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15.
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50.
Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–5.
Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49:515–26.
Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–20.
Pinkas DM, Sanvitale CE, Bufton JC, Sorrell FJ, Solcan N, Chalk R, et al. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem J. 2017;474:3747–61.
Dubiel W, Dubiel D, Wolf DA, Naumann M. Cullin 3-based ubiquitin ligases as master regulators of mammalian cell differentiation. Trends Biochem Sci. 2018;43:95–107.
Chen HY, Liu CC, Chen RH. Cul3-KLHL20 ubiquitin ligase: physiological functions, stress responses, and disease implications. Cell Div. 2016;11:5.
Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75.
Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–38.
McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–U1285.
Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–87.
Ha S, Sohn IJ, Kim N, Sim HJ, Cheon KA. Characteristics of brains in autism spectrum disorder: structure, function and connectivity across the lifespan. Exp Neurobiol. 2015;24:273–84.
Hernandez LM, Rudie JD, Green SA, Bookheimer S, Dapretto M. Neural signatures of autism spectrum disorders: insights into brain network dynamics. Neuropsychopharmacology. 2015;40:171–89.
Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, et al. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–19.
Schubert D, Martens GJ, Kolk SM. Molecular underpinnings of prefrontal cortex development in rodents provide insights into the etiology of neurodevelopmental disorders. Mol Psychiatry. 2015;20:795–809.
Shepherd GM. Corticostriatal connectivity and its role in disease. Nat Rev Neurosci. 2013;14:278–91.
McEvoy JD, Kossatz U, Malek N, Singer JD. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol Cell Biol. 2007;27:3651–66.
Gulisano M, Broccoli V, Pardini C, Boncinelli E. Emx1 and Emx2 show different patterns of expression during proliferation and differentiation of the developing cerebral cortex in the mouse. Eur J Neurosci. 1996;8:1037–50.
Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–14.
Perry W, Minassian A, Lopez B, Maron L, Lincoln A. Sensorimotor gating deficits in adults with autism. Biol Psychiatry. 2007;61:482–6.
Brumback AC, Ellwood IT, Kjaerby C, Iafrati J, Robinson S, Lee AT, et al. Identifying specific prefrontal neurons that contribute to autism-associated abnormalities in physiology and social behavior. Mol Psychiatry. 2017;23:2078–89.
Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 2015;11:1400–13.
Tebartz van Elst L, Maier S, Fangmeier T, Endres D, Mueller GT, Nickel K, et al. Disturbed cingulate glutamate metabolism in adults with high-functioning autism spectrum disorder: evidence in support of the excitatory/inhibitory imbalance hypothesis. Mol Psychiatry. 2014;19:1314–25.
Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007.
Ajram LA, Horder J, Mendez MA, Galanopoulos A, Brennan LP, Wichers RH, et al. Shifting brain inhibitory balance and connectivity of the prefrontal cortex of adults with autism spectrum disorder. Transl Psychiatry. 2017;7:e1137.
Fuccillo MV. Striatal circuits as a common node for autism pathophysiology. Front Neurosci. 2016;10:27.
Burguiere E, Monteiro P, Mallet L, Feng G, Graybiel AM. Striatal circuits, habits, and implications for obsessive-compulsive disorder. Curr Opin Neurobiol. 2015;30:59–65.
Langen M, Bos D, Noordermeer SD, Nederveen H, van Engeland H, Durston S. Changes in the development of striatum are involved in repetitive behavior in autism. Biol Psychiatry. 2014;76:405–11.
Kreitzer AC, Malenka RC. Endocannabinoid-mediated rescue of striatal LTD and motor deficits in Parkinson’s disease models. Nature. 2007;445:643–7.
Cepeda C, Andre VM, Yamazaki I, Wu N, Kleiman-Weiner M, Levine MS. Differential electrophysiological properties of dopamine D1 and D2 receptor-containing striatal medium-sized spiny neurons. Eur J Neurosci. 2008;27:671–82.
Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–55.
Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–26.
Yan Z, Kim E, Datta D, Lewis DA, Soderling SH. Synaptic actin dysregulation, a convergent mechanism of mental disorders? J Neurosci. 2016;36:11411–7.
Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10:927–38.
Quassollo G, Wojnacki J, Salas DA, Gastaldi L, Marzolo MP, Conde C, et al. A RhoA signaling pathway regulates dendritic golgi outpost formation. Curr Biol. 2015;25:971–82.
Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, et al. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 2015;85:742–54.
Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, et al. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch Gen Psychiatry. 2012;69:314–24.
Peserico A, Germani A, Sanese P, Barbosa AJ, Di Virgilio V, Fittipaldi R, et al. A SMYD3 small-molecule inhibitor impairing cancer cell growth. J Cell Physiol. 2015;230:2447–60.
Amodio DM, Frith CD. Meeting of minds: the medial frontal cortex and social cognition. Nat Rev Neurosci. 2006;7:268–77.
Qin L, Ma K, Wang ZJ, Hu Z, Matas E, Wei J, et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci. 2018;21:564–75.
Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–42.
Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–5.
Wang W, Rein B, Zhang F, Tan T, Zhong P, Qin LY, et al. Chemogenetic activation of prefrontal cortex rescues synaptic and behavioral deficits in a mouse model of 16p11.2 deletion syndrome. J Neurosci. 2018;38:5939–48.
Escamilla CO, Filonova I, Walker AK, Xuan ZX, Holehonnur R, Espinosa F, et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. 2017;551:227–31.
McGourty CA, Akopian D, Walsh C, Gorur A, Werner A, Schekman R, et al. Regulation of the CUL3 ubiquitin ligase by a calcium-dependent co-adaptor. Cell. 2016;167:525–38 e514.
Acknowledgements
We thank Xiaoqing Chen for excellent technical support. We acknowledge the support of New York State Center of Excellence in Bioinformatics and Life Sciences and University at Buffalo. This work was supported by: Nancy Lurie Marks Family Foundation (Z.Y.), National Institutes of Health (MH112237 and MH108842) (Z.Y.), Brain and Behavior Foundation (24910) (M.R.) and National Center for Advancing Translational Sciences of the National Institutes of Health (KL2TR001413) (M.R.).
Author information
Authors and Affiliations
Contributions
M.R. performed the behavioral, immunocytochemical, anatomical, and some biochemical experiments, carried out bioinformatic analyses, and wrote the draft. T.T., W.W. and P.Z. performed electrophysiological experiments and analyzed data. X.W. and J.Q. carried out proteomic experiments and analyzed the data. Z.W., L.F., L.Q. and K.M. participated in the biochemical, immunocytochemical, anatomical, and behavioral experiments. Z.Y. designed the experiments, supervised the project, and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Rapanelli, M., Tan, T., Wang, W. et al. Behavioral, circuitry, and molecular aberrations by region-specific deficiency of the high-risk autism gene Cul3. Mol Psychiatry 26, 1491–1504 (2021). https://doi.org/10.1038/s41380-019-0498-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0498-x
This article is cited by
-
A prefrontal-thalamic circuit encodes social information for social recognition
Nature Communications (2024)
-
Increased gene dosage of RFWD2 causes autistic-like behaviors and aberrant synaptic formation and function in mice
Molecular Psychiatry (2024)
-
Neuronal knockdown of Cullin3 as a Drosophila model of autism spectrum disorder
Scientific Reports (2024)
-
Severity of Autism Spectrum Disorder Symptoms Associated with de novo Variants and Pregnancy-Induced Hypertension
Journal of Autism and Developmental Disorders (2024)
-
Inhibition of histone methyltransferase Smyd3 rescues NMDAR and cognitive deficits in a tauopathy mouse model
Nature Communications (2023)