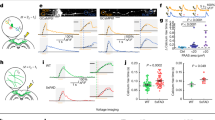
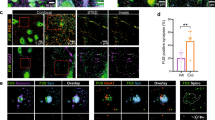
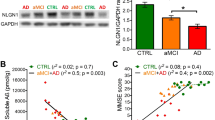
Abstract
Postsynaptic trafficking plays a key role in regulating synapse structure and function. While spiny excitatory synapses can be stable throughout adult life, their morphology and function is impaired in Alzheimer’s disease (AD). However, little is known about how AD risk genes impact synaptic function. Here we used structured superresolution illumination microscopy (SIM) to study the late-onset Alzheimer’s disease (LOAD) risk factor BIN1, and show that this protein is abundant in postsynaptic compartments, including spines. While postsynaptic Bin1 shows colocalization with clathrin, a major endocytic protein, it also colocalizes with the small GTPases Rab11 and Arf6, components of the exocytic pathway. Bin1 participates in protein complexes with Arf6 and GluA1, and manipulations of Bin1 lead to changes in spine morphology, AMPA receptor surface expression and trafficking, and AMPA receptor-mediated synaptic transmission. Our data provide new insights into the mesoscale architecture of postsynaptic trafficking compartments and their regulation by a major LOAD risk factor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol. 2012;22:461–9. https://doi.org/10.1016/j.conb.2011.12.006.
Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–93. https://doi.org/10.1038/nn.2741.
van der Sluijs P, Hoogenraad CC. New insights in endosomal dynamics and AMPA receptor trafficking. Semin Cell Dev Biol. 2011;22:499–505. https://doi.org/10.1016/j.semcdb.2011.06.008.
Moore FB, Baleja JD. Molecular remodeling mechanisms of the neural somatodendritic compartment. Biochim Biophys Acta. 2012;1823:1720–30. https://doi.org/10.1016/j.bbamcr.2012.06.006.
Hsu VW, Bai M, Li J. Getting active: protein sorting in endocytic recycling. Nat Rev Mol Cell Biol. 2012;13:323–8. https://doi.org/10.1038/nrm3332.
DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol. 1990;27:457–64. https://doi.org/10.1002/ana.410270502.
Bertram L, Tanzi RE. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–78. https://doi.org/10.1038/nrn2494.
Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. https://doi.org/10.1016/j.biopsych.2014.05.006.
Zhu JB, Tan CC, Tan L, Yu JT. State of play in Alzheimer’s disease genetics. J Alzheimers Dis. 2017;58:631–59. https://doi.org/10.3233/JAD-170062.
Tan MS, Yu JT, Tan L. Bridging integrator 1 (BIN1): form, function, and Alzheimer’s disease. Trends Mol Med. 2013;19:594–603. https://doi.org/10.1016/j.molmed.2013.06.004.
Prokic I, Cowling BS, Laporte J. Amphiphysin 2 (BIN1) in physiology and diseases. J Mol Med. 2014;92:453–63. https://doi.org/10.1007/s00109-014-1138-1.
Picas L, et al. BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat Commun. 2014;5:5647 https://doi.org/10.1038/ncomms6647.
Drager NM, et al. Bin1 directly remodels actin dynamics through its BAR domain. EMBO Rep. 2017;18:2051–66. https://doi.org/10.15252/embr.201744137.
Nakajo A, et al. EHBP1L1 coordinates Rab8 and Bin1 to regulate apical-directed transport in polarized epithelial cells. J Cell Biol. 2016;212:297–306. https://doi.org/10.1083/jcb.201508086.
Pant S, et al. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–410. https://doi.org/10.1038/ncb1986.
Bayes A, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. https://doi.org/10.1038/nn.2719.
Grossmann, AH et al. The small GTPase ARF6 regulates protein trafficking to control cellular function during development and in disease. Small GTPases. https://doi.org/10.1080/21541248.2016.1259710 (2016).
Hongu T, Kanaho Y. Activation machinery of the small GTPase Arf6. Adv Biol Regul. 2014;54:59–66. https://doi.org/10.1016/j.jbior.2013.09.014.
Johnson DL, Wayt J, Wilson JM, Donaldson JG. Arf6 and Rab22 mediate T cell conjugate formation by regulating clathrin-independent endosomal membrane trafficking. J Cell Sci. 2017;130:2405–15. https://doi.org/10.1242/jcs.200477.
Harris, KM & Weinberg, RJ Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a005587 (2012).
Dhar SS, Liang HL, Wong-Riley MT. Nuclear respiratory factor 1 co-regulates AMPA glutamate receptor subunit 2 and cytochrome c oxidase: tight coupling of glutamatergic transmission and energy metabolism in neurons. J Neurochem. 2009;108:1595–606. https://doi.org/10.1111/j.1471-4159.2009.05929.x
Caplan S, et al. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–67. https://doi.org/10.1093/emboj/21.11.2557
Posey AD Jr., et al. EHD1 mediates vesicle trafficking required for normal muscle growth and transverse tubule development. Dev Biol. 2014;387:179–90. 10.1016/j.ydbio.2014.01.004
Oku Y, Huganir RL. AGAP3 and Arf6 regulate trafficking of AMPA receptors and synaptic plasticity. J Neurosci. 2013;33:12586–98. https://doi.org/10.1523/JNEUROSCI.0341-13.2013
Scholz R, et al. AMPA receptor signaling through BRAG2 and Arf6 critical for long-term synaptic depression. Neuron. 2010;66:768–80. https://doi.org/10.1016/j.neuron.2010.05.003.
Penzes P, Vanleeuwen JE. Impaired regulation of synaptic actin cytoskeleton in Alzheimer’s disease. Brain Res Rev. 2011;67:184–92. https://doi.org/10.1016/j.brainresrev.2011.01.003.
Karch CM, Cruchaga C, Goate AM. Alzheimer’s disease genetics: from the bench to the clinic. Neuron. 2014;83:11–26. https://doi.org/10.1016/j.neuron.2014.05.041.
Glennon EB, et al. BIN1 is decreased in sporadic but not familial Alzheimer’s disease or in aging. PLoS One. 2013;8:e78806 https://doi.org/10.1371/journal.pone.0078806.
Adams SL, Tilton K, Kozubek JA, Seshadri S, Delalle I. Subcellular changes in bridging integrator 1 protein expression in the cerebral cortex during the progression of Alzheimer disease pathology. J Neuropathol Exp Neurol. 2016;75:779–90. https://doi.org/10.1093/jnen/nlw056.
Holler CJ, et al. Bridging integrator 1 (BIN1) protein expression increases in the Alzheimer’s disease brain and correlates with neurofibrillary tangle pathology. J Alzheimers Dis. 2014;42:1221–7. https://doi.org/10.3233/JAD-132450.
Lee E, et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–6. https://doi.org/10.1126/science.1071362.
Copits BA, Swanson GT. Kainate receptor post-translational modifications differentially regulate association with 4.1N to control activity-dependent receptor endocytosis. J Biol Chem. 2013;288:8952–65. https://doi.org/10.1074/jbc.M112.440719.
Smith KR, et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron. 2014;84:399–415. https://doi.org/10.1016/j.neuron.2014.10.010.
Burette AC, et al. Organization of TNIK in dendritic spines. J Comp Neurol. 2015;523:1913–24. https://doi.org/10.1002/cne.23770.
Kopec CD, Li B, Wei W, Boehm J, Malinow R. Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J Neurosci. 2006;26:2000–9. https://doi.org/10.1523/JNEUROSCI.3918-05.2006.
Acknowledgements
This work was supported by: R01s MH097216, MH107182, and MH097216 and R56 AG063433 to PP, German Research Foundation (DFG) Postdoctoral Research Fellowship SCHU2710/1-1 to BS, NS064091 MM, and NS039444 to RJW. Imaging work was partly performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Structured illumination microscopy was performed on a Nikon N-SIM system, purchased through the support of NIH 1S10OD016342-01. We thank Joshua Rappoport, Constadina Arvanitis, and Teng Leong Chew for assistance with imaging and analysis. All experiments involving animals were performed according to the Institutional Animal Care and Use Committee of NU.
Author information
Authors and Affiliations
Contributions
BS performed and analyzed confocal and SIM imaging experiments, some biochemistry, led the project and wrote the paper. DPB performed FRAP experiments, Arf6 activation assays, GluA1 surface expression experiments, PLA experiments, some biochemistry, data analysis and manuscript writing. KJK performed some SIM imaging. SY performed Bin1 developmental expression experiments. KM performed GluA1 surface expression experiments and FRAP experiments. KH provided general help for all experiments. CJK performed electrophysiology experiments. MDMS, MF, KRS performed biochemistry experiments. JMFP and RG performed molecular biology. MM supervised electrophysiology and advised on the project. ACB performed electron microscopy experiments. JR assisted with SIM imaging studies and data analysis. RJW supervised the electron microscopy experiments. PP supervised the project and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Schürmann, B., Bermingham, D.P., Kopeikina, K.J. et al. A novel role for the late-onset Alzheimer’s disease (LOAD)-associated protein Bin1 in regulating postsynaptic trafficking and glutamatergic signaling. Mol Psychiatry 25, 2000–2016 (2020). https://doi.org/10.1038/s41380-019-0407-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0407-3
This article is cited by
-
The Alzheimer’s disease risk gene BIN1 regulates activity-dependent gene expression in human-induced glutamatergic neurons
Molecular Psychiatry (2024)
-
The Alzheimer susceptibility gene BIN1 induces isoform-dependent neurotoxicity through early endosome defects
Acta Neuropathologica Communications (2022)
-
Synaptic tau: A pathological or physiological phenomenon?
Acta Neuropathologica Communications (2021)
-
Vezatin regulates seizures by controlling AMPAR-mediated synaptic activity
Cell Death & Disease (2021)