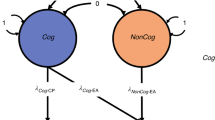
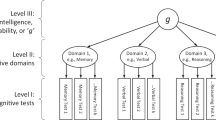
Abstract
Recent advances in genomics are producing powerful DNA predictors of complex traits, especially cognitive abilities. Here, we leveraged summary statistics from the most recent genome-wide association studies of intelligence and educational attainment, with highly genetically correlated traits, to build prediction models of general cognitive ability and educational achievement. To this end, we compared the performances of multi-trait genomic and polygenic scoring methods. In a representative UK sample of 7,026 children at ages 12 and 16, we show that we can now predict up to 11% of the variance in intelligence and 16% in educational achievement. We also show that predictive power increases from age 12 to age 16 and that genomic predictions do not differ for girls and boys. We found that multi-trait genomic methods were effective in boosting predictive power. Prediction accuracy varied across polygenic score approaches, however results were similar for different multi-trait and polygenic score methods. We discuss general caveats of multi-trait methods and polygenic score prediction, and conclude that polygenic scores for educational attainment and intelligence are currently the most powerful predictors in the behavioural sciences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Plomin R, von Stumm S. The new genetics of intelligence. Nat Rev Genet. 2018;19:148–59.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nat Rev Genet. 2017;18:117–27.
Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–37.
Maier RM, Zhu Z, Lee SH, Trzaskowski M, Ruderfer DM, Stahl EA, et al. Improving genetic prediction by leveraging genetic correlations among human diseases and traits. Nat Commun. 2018;9:989.
Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion MJ, et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2014;19:253–8.
Butcher LM, Davis OS, Craig IW, Plomin R. Genome-wide quantitative trait locus association scan of general cognitive ability using pooled DNA and 500K single nucleotide polymorphism microarrays. Genes Brain Behav. 2008;7:435–46.
Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53,949). Mol Psychiatry. 2015;20:183–92.
Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N= 112,151). Mol Psychiatry. 2016;21:758–67.
Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005.
Plomin R, Hill L, Craig IW, McGuffin P, Purcell S, Sham P, et al. A genome-wide scan of 1842 DNA markers for allelic associations with general cognitive ability: a five-stage design using DNA pooling and extreme selected groups. Behav Genet. 2001;31:497–509.
Trampush JW, Yang ML, Yu J, Knowles E, Davies G, Liewald DC, et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry. 2017;22:336–45.
Sniekers S, Stringer S, Watanabe K, Jansen PR, Coleman JRI, Krapohl E, et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet. 2017;49:1107–12.
Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–9.
Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–71.
Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–42.
Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–21
Krapohl E, Plomin R. Genetic link between family socioeconomic status and children’s educational achievement estimated from genome-wide SNPs. Mol Psychiatry. 2016;21:437–43.
Selzam S, Krapohl E, von Stumm S, O’Reilly PF, Rimfeld K, Kovas Y, et al. Predicting educational achievement from DNA. Mol Psychiatry. 2018;23:161.
Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci USA. 2014;111:13790–4.
Krapohl E, Patel H, Newhouse S, Curtis CJ, von Stumm S, Dale PS, et al. Multi-polygenic score approach to trait prediction. Mol Psychiatry. 2018;23:1368–74.
Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31:1466–8.
Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–92.
Mak TSH, Porsch RM, Choi SW, Zhou X, Sham PC. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 2017;41:469–80.
Hill WD, Hagenaars SP, Marioni RE, Harris SE, Liewald DCM, Davies G, et al. Molecular genetic contributions to social deprivation and household income in UK biobank. Curr Biol. 2016;26:3083–9.
Seed C Hail: An Open-Source Framework for Scalable Genetic Data. 2017.
Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD et al. Genomic SEM provides insights into the multivariate genetic architecture of complex traits. bioRxiv 2018.
Haworth CM, Davis OS, Plomin R. Twins Early Development Study (TEDS): a genetically sensitive investigation of cognitive and behavioral development from childhood to young adulthood. Twin Res Hum Genet. 2013;16:117–25.
Oliver BR, Plomin R. Twins’ Early Development Study (TEDS): a multivariate, longitudinal genetic investigation of language, cognition and behavior problems from childhood through adolescence. Twin Res Hum Genet. 2007;10:96–105.
Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. s361-363
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Krapohl E, Euesden J, Zabaneh D, Pingault JB, Rimfeld K, von Stumm S, et al. Phenome-wide analysis of genome-wide polygenic scores. Mol Psychiatry. 2015;21:1188.
Haworth CMA, Wright MJ, Luciano M, Martin NG, de Geus EJC, van Beijsterveldt CEM, et al. The heritability of general cognitive ability increases linearly from childhood to young adulthood. Mol Psychiatry. 2010;15:1112–20.
Plomin R. Blueprint: how DNA makes us who we are. London: Allen Lane/Penguing Press; 2018.
Fletcher JM, Lehrer SF. Genetic lotteries within families. J Health Econ. 2011;30:647–59.
Pingault J-B, O’Reilly PF, Schoeler T, Ploubidis GB, Rijsdijk F, Dudbridge F. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet. 2018;19:566–80.
Belsky DW, Domingue BW, Wedow R, Arseneault L, Boardman JD, Caspi A, et al. Genetic analysis of social-class mobility in five longitudinal studies. Proc Natl Acad Sci. 2018;115:E7275–E7284.
Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, et al. The nature of nurture: effects of parental genotypes. Science. 2018;359:424–8.
Plomin R, Bergeman CS. The nature of nurture: genetic influence on “environmental” measures. Behav Brain Sci. 2011;14:373–86.
Plomin R. Genetics and experience: the interplay between nature and nurture. Thousand Oaks, CA: Sage Publications; 1994.
Krapohl E, Plomin R. Genetic link between family socioeconomic status and children’s educational achievement estimated from genome-wide SNPs. Mol Psychiatry. 2015;21:437.
Acknowledgements
We gratefully acknowledge the ongoing contribution of the participants in the Twins Early Development Study (TEDS) and their families. TEDS is supported by a programme grant to RP from the UK Medical Research Council (MR/M021475/1 and previously G0901245), with additional support from the US National Institutes of Health (AG046938). The research leading to these results has also received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/grant agreement n° 602768 and ERC grant agreement n° 295366. RP is supported by a Medical Research Council Professorship award (G19/2). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 721567.
Author information
Authors and Affiliations
Contributions
AGA and RP conceived and designed the study. AGA analysed and interpreted the data. SS performed quality control of genotype data. AGA and RP wrote the manuscript. SS, KR, SvS and JBP contributed to and critically reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Allegrini, A.G., Selzam, S., Rimfeld, K. et al. Genomic prediction of cognitive traits in childhood and adolescence. Mol Psychiatry 24, 819–827 (2019). https://doi.org/10.1038/s41380-019-0394-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0394-4
This article is cited by
-
Utility of polygenic scores across diverse diseases in a hospital cohort for predictive modeling
Nature Communications (2024)
-
Infrastructuring Educational Genomics: Associations, Architectures, and Apparatuses
Postdigital Science and Education (2024)
-
Behavioural genetics methods
Nature Reviews Methods Primers (2023)
-
Multibreed genomic prediction using summary statistics and a breed-origin-of-alleles approach
Heredity (2023)
-
Celebrating a Century of Research in Behavioral Genetics
Behavior Genetics (2023)