Abstract
The cognitive mechanisms underlying attention-deficit hyperactivity disorder (ADHD), a highly heritable disorder with an array of candidate genes and unclear genetic architecture, remain poorly understood. We previously demonstrated that mice overexpressing CK1δ (CK1δ OE) in the forebrain show hyperactivity and ADHD-like pharmacological responses to d-amphetamine. Here, we demonstrate that CK1δ OE mice exhibit impaired visual attention and a lack of d-amphetamine-induced place preference, indicating a disruption of the dopamine-dependent reward pathway. We also demonstrate the presence of abnormalities in the frontostriatal circuitry, differences in synaptic ultra-structures by electron microscopy, as well as electrophysiological perturbations of both glutamatergic and GABAergic transmission, as observed by altered frequency and amplitude of mEPSCs and mIPSCs. Furthermore, gene expression profiling by next-generation sequencing alone, or in combination with bacTRAP technology to study specifically Drd1a versus Drd2 medium spiny neurons, revealed that developmental CK1δ OE alters transcriptional homeostasis in the striatum, including specific alterations in Drd1a versus Drd2 neurons. These results led us to perform a fine molecular characterization of targeted gene networks and pathway analysis. Importantly, a large fraction of 92 genes identified by GWAS studies as associated with ADHD in humans are significantly altered in our mouse model. The multiple abnormalities described here might be responsible for synaptic alterations and lead to complex behavioral abnormalities. Collectively, CK1δ OE mice share characteristics typically associated with ADHD and should represent a valuable model to investigate the disease in vivo.
Similar content being viewed by others
Introduction
Attention-deficit hyperactivity disorder (ADHD), characterized by excessive levels of inattention, hyperactivity, and impulsivity, is prevalent worldwide, affecting about 5.3% of school-age children, and persisting into adulthood in about two-thirds of such children [1,2,3]. ADHD is a developmental disorder and pathophysiological abnormalities of the frontostriatal network have been observed [4]. Large twin studies have revealed that ADHD is highly heritable [5], and recent work by the Psychiatric Genomics Consortium using genome-wide association studies (GWAS) has demonstrated that the disorder is polygenic [6, 7], with clear evidence of shared genetic risk at individual loci. Nevertheless, the diagnostic etiology and the genetic–neurophysiological mechanisms underlying the disorder still remain difficult to pinpoint.
Using magnetic resonance imaging (MRI), abnormal activation in the prefrontal cortex and striatum has been observed in ADHD patients [8,9,10]. Precise topological measurements using functional MRI showed that activation of the visual attention network is significantly reduced in ADHD children, indicating non-coordinated functional activity from the cingulate cortex to other brain regions [11]. It has been reported that impaired interactions between control and reward pathways exist in ADHD brains underlying attention and motivation deficits in the patients [12]. Indeed, abnormal patterns of activity in dopamine neurotransmission have been implicated in the pathophysiology of ADHD [13,14,15], and positron emission tomography studies in ADHD patients have revealed altered changes in dopamine transmission in the reward-motivation pathway [16, 17]. From a signaling point of view, an imbalance of dopamine neurotransmission in the nigrostriatal pathway has been linked to ADHD [18,19,20]. Methylphenidate, mainly acting on dopamine transmission, is still the first-line pharmacological treatment of ADHD [21]. Remarkably, ADHD has also been associated with a series of other psychiatric conditions where dopamine is also involved, such as major depression and bipolar disorder and autism [22, 23].
Animal models provide testable and valuable tools for deep understanding of the etiology, pathology, and therapeutic treatment of diseases. The behavioral performance of mice lacking the dopamine transporter, at baseline and in response to stimulant drugs, firmly support dysregulation of key neurotransmitters in ADHD [24, 25]. However, dopamine neurotransmission can be impacted by many factors. In the basal ganglia, DARPP-32 (dopamine- and cAMP-regulated phosphoprotein, mw 32 kDa) is a key protein that integrates synaptic input signals from various origins including the dopaminergic system [24]. CK1 regulates the state of DARPP-32 phosphorylation, influencing neuronal function through site-specific phosphorylation. CK1δ is a major CK1 isoform and is enriched in the brain [26]. In the central nervous system, CK1δ is involved in a variety of physiological and pathological processes [27,28,29]. To assess the role of CK1δ in the regulation of dopamine signaling, we previously generated a transgenic mouse line overexpressing CK1δ (CK1δ OE) in the forebrain in a temporally controllable manner. CK1δ OE mice exhibited hyperactive locomotion, impulsivity, and lower nesting capability. Behavioral performance normalized when the OE mice were administrated amphetamine or methylphenidate, effective drugs currently used in patients with ADHD [30].
Our previous work indicates that CK1δ OE plays an important role in perturbation of dopaminergic signaling, implicating mechanisms underlying ADHD [30]. Here, we specifically assessed attention deficit in CK1δ OE mice and studied the function of brain rewarding circuits by conducting a series of behavioral studies. To further investigate the mechanisms underlying these behavioral abnormalities, we then examined the structural changes, neuronal morphology, and synaptic ultrastructural differences in the frontostriatal areas of the CK1δ OE mice. We next investigated the impact of these modifications on electrophysiological recordings and demonstrated electrophysiological perturbances of both glutamatergic and GABAergic transmission by analyzing frequency and kinetics of miniature excitatory postsynaptic currents (mEPSCs) and miniature inhibitory postsynaptic currents (mIPSCs). Furthermore, we analyzed transcriptional gene expression in brain areas associated with these complex abnormalities. To further investigate ADHD-related abnormalities, and address the still debatable roles of dopamine receptor 1 (Drd1a) versus dopamine receptor 2 (Drd2) in medium spiny neurons (MSNs), we investigated the possibility that the pathways of Drd1a and Drd2 MSNs might be affected differentially by CK1δ OE. We then obtained the mice with the combination of bacterial artificial chromosome and translating ribosome affinity purification (bacTRAP) and performed RNA profiling on four different neuronal populations, showing some clear differences between cell types for the two genotypes in the striatum. Finally, we explored a possible relationship between CK1δ OE-induced gene expression and the ADHD risk genes, over 90 human genes associated with ADHD through GWAS studies.
Our findings demonstrate that the temporal and regional inducibility of CK1δ OE in our mouse model provides a unique tool to elucidate in vivo the mechanisms underlying ADHD. Together, this study demonstrates the importance of CK1δ regulation in the pathophysiology of ADHD, reinforces the importance of the dopaminergic system, involving both classes of MSNs, and provides a pharmacological framework to alleviate ADHD symptoms. Current animal models recapitulating the behavioral, physiological, genetic, and mechanistic complexity of ADHD are lacking. The comprehensive investigation described herein suggests that CK1δ OE is a good model of this highly prevalent disorder.
Materials and methods
Animals used and behavioral tests
Mouse procedures were in accordance with the National Institutes of Health guidelines and approved by the Institutional Animal Care and Use Committees at The Rockefeller University. Male mice were used in all behavioral tests, and both males and females were processed for tissue preparations.
The transgenic mouse line with inducible overexpression of CK1δ in the forebrain was generated by crossing tetO-CK1δ-positive mice with CaMKIIα-tTA-positive mice [30]. CK1δ/tTA/Drd1a/EGFP-L10a and CK1δ/tTA/Drd2/EGFP-L10a bacTRAP transgenic mice were generated on a mixed genetic background (The Jackson Laboratory, Bar Harbor, Maine) by breeding the mice carrying CK1δ+/tTA+ with Drd1a/EGFP-L10a+ and Drd2/EGFP-L10a+. bacTRAP lines expressing the transgene EGFP-L10a under the promoter of Drd1a and Drd2 were described elsewhere [31].
The locomotor activities in both sexes were evaluated in an open field test (60 min). Male CK1δ OE showed hyper-locomotion at 1 month and 16 months of age.
Female CK1δ OE were similarly hyperactive in young adulthood (3 months of age), but not when they were older at 6–7 and 16 months of age. Since the behavioral effect of the CK1δ OE was more robust and longer lasting in males than in females, males were selected for the subsequent behavioral tests, such as for 3-Choice Serial Reaction Time task (3CSRT), the complexity of which required multiple months of training and testing. When young animals were used (<3 months of age) both males and females were used for biochemical assays.
The 3CSRT test was used to assess attentional function [32, 33]. Prior to the test, mice were food restricted to reduce their body weights by around 15%. The mice were then trained (20 min daily) to collect one food pellet dispensed into the magazine (Med Associates, St. Albans, VT) and to nose-poke in any one of three lit apertures (stimulus light went from constant to random) in order to obtain one food pellet 20 min daily. The training phase was completed when mice obtained 40 food rewards and at least 50% of correct responses. To begin the first trial, mice had to nose-poke the randomly illuminated aperture (lit for 60 s) with 5 s delay between trials. A correct response (i.e. a nose-poke in the illuminated aperture) resulted in delivery of a food pellet and then initiated the next trial. Response accuracy (i.e. the percentage of correct responses/total trials) was recorded for each session, and mice advanced to the next stage of the procedure when they performed >50 trials per session and achieved >80% correct actions. The stimulus duration decreased progressively at each stage from 60 to 32, 16, 8, 4, 2, and 1 s. Variables measured in this experiment included number of days required to reach the above criterion for each stimulus duration, food magazine entries, accuracy, and timeout counts. An 8 s timeout occurred when any error was made, including anticipatory errors (nose-poking prior to delivery of the visual stimulus), commission error (nose-poking in either of the non-illuminated apertures), and omission error (no nose-poke during the stimulus or the subsequent 5-s holding period).
Amphetamine-based reward testing was performed using an adapted version of existing protocols [34,35,36] and conducted from 2 to 5 p.m. The first three days, mice were individually placed in the central chamber of the three chambered apparatus for 30 min daily and allowed to freely explore all three chambers during the test. On Days 4 and 5 the mice received d-amphetamine (2 mg/kg via i.p.) and were immediately transferred into the center chamber of the CPP apparatus where they were given access to all three chambers for 30 min. On Day 6, all mice received saline (0.9% of NaCl) and were immediately placed in the center chamber, where they were again given access to the three chambers for 30 min. On Days 7 and 8, the mice were immediately confined into the black chamber (drug-paired) for 30 min immediately after receiving d-amphetamine (2 mg/kg via i.p.). Thereafter, the mice were kept drug free. On Day 9, 24 h after amphetamine withdrawal, mice were placed into the central chamber with all doors opened for 30 min. Six days later (see Day 9), the procedure was repeated. The entries to each chamber and the time spent inside each chamber were recorded daily.
Golgi staining and striatal volume measurements
The Golgi staining was performed as indicated in the user manual, PK401, from FD NeuroTechnologies Inc (Ellicott City, MD, USA). See also Supplementary Material for details. The striatum of each section (circled areas) were measured using ImageJ (NIH). The striatal volumes were calculated as the sum of all striatal section areas corresponding to a single brain hemisphere and multiplied by 100 to take into account the thickness of the sections.
Transmission electron microscopy
Brains were perfused 2–3 min with saline and followed by fixation, containing 2% paraformaldehyde and 2.5% glutaraldehyde in 0.75 M sodium cacodylate buffer (pH 7.4) at 25 °C for 5 min. The brains were kept in the fixative solution at 4 °C overnight. Vibratome sections were generated, washed, post-fixed with 1% osmium tetra-oxide for 1 h, underwent en bloc staining with 2% uranyl acetate for30 min, dehydrated by a graded series of ethanol, removed ethanol with propylene oxide, infiltrated with a resin (EMBed 812; Electron Microscope Sciences, Hatfield, PA) and embedded with the resin. After polymerization at 60 °C for 48 h, ultra-thin sections underwent post-staining with 2% uranyl acetate and 1% lead citrate and were examined in the electron microscope (100CX JEOL, Tokyo, Japan) with the digital imaging system (XR41-C; Advantage Microscopy Technology (AMT) Corp., Denver, MA).
Electrophysiology
Male mice, 4–5-weeks old, were sacrificed by a Euthanex SmartboxTM Euthanasia System (Euthanex, Palmer, PA, USA) and decapitation. Brains were removed quickly, placed in ice-cold artificial cerebrospinal fluid, coronal brain slices (300 μm thickness) were prepared using a VT-1000S Vibratome (Leica Microsystems Inc., Buffalo Grove, IL, USA) and allowed to recover for 1 h at 35 °C.
Electrophysiological recordings and data analysis were performed as previously described [37, 38]. Striatum-containing slices were placed in a perfusion chamber attached to the fixed stage of an upright BX51WI microscope (Olympus, Japan) and submerged in continuously flowing oxygenated recording solution. Neurons were visualized with a ×40 water immersion lens and illuminated with near infrared (IR) light. Striatal MSNs were identified based on their size, morphology, and electrophysiological characteristics (resting membrane potential and spike discharge). Whole-cell, voltage-clamp recordings from striatal MSNs were performed with a Multiclamp 700B/Digidata1440A system (Molecular Devices, Sunnyvale, CA, USA). Neurons were voltage-clamped at −70 mV and allowed to reach a stable baseline (5 min) before recording. To isolate and record mEPSCs, 10 μM bicuculline (Tocris Bioscience, Ellisville, MO, USA) to block GABA receptors and 0.5 μM tetrodotoxin (TTX; Tocris Bioscience) to block sodium channels were added to the extracellular recording solution. To record mIPSCs, 50 μM D-(2 R)-amino-5-phosphonovaleric acid (D-APV; Tocris Bioscience), 20 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX; Tocris Bioscience), and 0.5 μM TTX were added to the recording solution to block NMDA glutamate receptors, AMPA/kainate glutamate receptors, and sodium channels, respectively.
All data were acquired with Clampex 10.3 (pClamp; Molecular Devices, Sunnyvale, CA). Amplitude, frequency, 10–90% rise time, and decay time were analyzed with MiniAnalysis (Synaptosoft Inc., Fort Lee, NJ, USA) and Prism 6 (GraphPad Software, La Jolla, CA, USA) software. The mEPSC decay time was fitted with a single exponential curve. The mIPSC decay time was fitted with a bi-exponential curve (A1 × exp(−tτ1) + A2 × exp(−t/τ2)) and the weighted decay constant (τw = (A1 × τ1 + A2 × τ2)/(A1 + A2)) was later computed. Unpaired Student’s t-test was used for statistical comparisons of amplitudes, frequencies, rise times, and decay times. All data are presented as mean ± s.e.m.
See also Supplementary Material for details.
RNA sequencing
Total RNAs from the frontal cortex and striatum of12-week-old mice, CK1δ+/tTA− and CK1δ+/tTA+, age- and sex-matched littermate controls (n = 3 or 4 per genotype), were extracted using the Trizol/Chloroform (Thermo Fisher, Carlsbad, CA). The extracted RNAs were precipitated in isopropanol and further purified using the Rneasy Micro Kit (Qiagen, Valencia, CA) with DNase digestion. The quality of purified RNAs were assessed using a Nanodrop 1000 spectrophotometer (ThermoScientific, Wilmington, DE) and a Bioanalyzer (Agilent Technologies, Santa Clara, CA).
For genomic arrays, purified mRNAs were amplified and processed using the Affymetrix two cycle cDNA Synthesis kit (Affymetrix, Santa Clara, CA). Mouse Genome 430 2.0 arrays were scanned using the GeneChip Scanner 3000 (Affymetrix, Santa Clara, CA) and globally scaled to 150 using the Affymetrix GeneChip Operating Software (GCOS v1.4). Three replicates were performed for each experiment. GeneChip CEL files were imported together into Genespring GX 7.3.1 (Agilent Technologies, Santa Clara, CA), processed with the GCRMA algorithm, and expression values on each chip were normalized to that chip’s 50th percentile. Gene changes (>2 folds) and significantly different (P values <0.05) between the two genotypes in the striatum and fronto-cortex, were selected.
For RNA sequence, the cDNAs were prepared with the TruSeq RNA Sample Preparation v2 kit (Illumina), using an input of 100 ng RNA for total striatal tissue and 25 ng RNA for bacTRAP samples from striatal either Drd1a or Drd2 neurons. Five hundred nanograms cDNA from each sample was fragmented on a Covaris S2 Focused Ultrasonicator using the operating conditions recommended by the manufacturer for a target fragment size of 200 bp. Libraries for RNA sequencing were prepared with the TruSeq RNA Sample Preparation v2 kit (Illumina), starting the manufacturer’s low-throughput protocol with the end-repair step. Subsequently, two libraries with different adapters were multiplexed for sequencing. The multiplexed samples were analyzed on an Illumina HiSeq 2000 sequencer using 100-bp single-end sequencing.
FPKM values for all genes in each sample were calculated with Cufflinks version 2.2.1. To analyze differential gene expression between samples (Supplementary Tables S1–S3), DESeq version 1.22.1 was used, applying the standard comparison mode. P-values were reported by DESeq, adjusted for multiple testing using the Benjamini–Hochberg procedure.
See also Supplementary Material for details and analysis.
Ingenuity pathway analysis
Data were analyzed, and functional networks were built, through the use of Ingenuity pathway analysis (IPA®; Qiagen Inc., https://www.qiagenbio informatics.com/products/ingenuitypathway-analysis). Briefly, RNAseq data were uploaded into Qiagen’s IPA system for basic analysis. Canonical pathways and gene networks relevant for diseases and functions were then evaluated through IPA based on pre-determined conditions (P-value <0.05 and fold change >1.5).
Heatmap analysis was used to illustrate the expression patterns of differentially expressed genes.
Statistical analysis
Parametric statistics were performed using StatView 5.0. Data are presented as means ± s.e.m. and were analyzed with one-way or two-way analysis of variance (ANOVA), or repeated measures two-way ANOVA with subject matching. Data were analyzed with Newman–Keuls post hoc analysis or Fisher’s protected least significant difference to compare differences between groups/within groups. Statistical significance was set and presented as *P < 0.05, **P < 0.005, and ***P < 0.001 genotype difference, and # P<0.05 difference in two conditions within one genotype.
Results
CK1δ OE mice show lower visual attention and impaired reward motivation
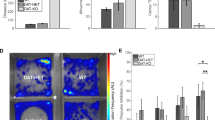
To further investigate the behavioral performance of CK1δ OE mice overexpressing CK1δ under the control of the CamKIIα-tTA promoter as a model of ADHD, we conducted the 3CSRT test to measure visual attention in the mice as follows. The stimulus duration (i.e. light) lasted for 60, 32, 16, 8, 4, 2, or 1 s depending on the testing phase. The criteria to pass each phase were set as 80% of correct actions and at least 50 completed trials. The number of CK1δ OE mice to reach criterion dropped quickly for the more difficult phases (8, 4, 2, 1 s), while 100% of control littermates passed most phases except the last one (1 s) where two animals failed (Fig. 1a). Additionally, even for the easier phases (60, 32, and 16 s), the CK1δ OE mice needed more days to pass each phases compared to controls (P = 0.031) (Fig. 1b). Furthermore, CK1δ OE mice checked the food magazine throughout the trial more often than control mice (Fig. 1c) (32″, F1,19 = 5.07, P = 0.036; 16″, F1,18 = 7.23, P = 0.015; 4″, F1,15 = 7.75, P = 0.014; 2″, F1,14 = 7.08, P = 0.019), indicative of a possible lowered inhibition, reminiscent of increased impulsivity associated with ADHD. The accuracy of CK1δ OE mice to respond to visual stimuli was significantly lower compared to control littermates, as measured by the average of correct trials divided by the total number of trials for each phase (Fig. 1d) (60″, F1,19 = 8.64, P = 0.008; 16″, F1,18 = 7.11, P = 0.016; 4″, F1,15 = 11.95, P = 0.0035; 2″, F1,14 = 48.96, P ≤ 0.0001; 1″, F1,9 = 23.52, P = 0.0009). In addition, CK1δ OE mice were subjected to more timeouts (penalties for wrong responses) compared to control mice (Fig. 1e) (16″, F1,18 = 9.87, P = 0.0056; 4″, F1,15 = 6.68, P = 0.021; 2″, F1,14 = 8.81, P = 0.0102).
CK1δ OE mice display lower visual attention and impairment in amphetamine-induced place preference compared to control littermates. a Number of mice to pass criterion in each phase. The duration of stimulus was variable, lasting for 60, 32, 16, 8, 4, 2, and 1 s. b Number of days needed to pass the first three phases (collapsed across phases). c Number of food magazine entries. d Accuracy to pass each phase (correct trials divided by the total number of trials), and e timeout counts. Statistical analysis: b Repeated measures two-way ANOVA with subject matching and fisher post hoc test, n = 10, *P < 0.05 genotype difference. c-e One-way ANOVA n = 10 each genotype, *P < 0.05 and ***P <0.001, genotype difference
Motivational deficits in ADHD patients may underlie a dysfunction of both attention and reward-motivation networks [12]. We use a d-amphetamine-based reward paradigm to study the rewarding effects of d-amphetamine (2 mg/kg) in our OE mouse model. A schematic representation of the procedure and the apparatus used for CPP is shown in Supplementary Figure S1a, b. In the first three days prior to drug injection, naive mice in both genotype groups did not show a significant difference in the time spent in the dark (right) chamber and in the number of entries into the dark chamber (Supplementary Figures S1c, d, marked d1–d3, pre-drug). However, on Day 9 control mice spent more time in the drug paired (dark) chamber compared to time spent in that side during the pre-drug days, as expected following amphetamine-induced place preference. Surprisingly, d-amphetamine exposure did not alter CK1δ OE mouse chamber preference (Supplementary Figures S1c, *F 1,18 = 6.32, P = 0.02; #F1,18 = 5.49, P = 0.031 difference in two test conditions within one group). Six days after withdrawing d-amphetamine (i.e. d15, post-drug), WT, but not CK1δ OE, mice still showed a tendency to spend more time in the drug-paired chamber (Supplementary Figures S1c).
CK1δ OE mouse brains harbor morphological and electrophysiological abnormalities
It has been reported that children with ADHD show a functional deficit in cortico-striatal circuitry [39, 40]. To investigate the origin of the ADHD-like phenotypes observed in CK1δ OE mice, we performed histological experiments using Golgi staining and immunohistochemistry, focusing on the cortico-striatal regions. CK1δ OE mice have shorter neuronal dendritic branches and lower neuronal density in the cingulate cortex compared to control littermates (Fig. 2a and Supplementary Figure S2a). This result was confirmed quantitatively by counting the total number of neurons (NeuN positive) in the area of interest (Fig. 2b and Supplementary Figure S2b) (n = 5 sections, 3 mice for each genotype F1, 6 = 12.25; P = 0.012).
CK1δ OE mouse brains harbor morphological and electrophysiological abnormalities. a Coronal sections of Golgi stained brains (×10). CK1δ OE mice harbor shorter neuronal dendrites in the cingulate cortex compared to control littermates. b Bar graph shows the statistical result of NeuN cell counts in the cingulate ctx (one-way ANOVA, n = 5; 3 brains for each genotype), *P < 0.05. c Statistical analysis of the striatal volumes (one-way ANOVA n = 6 for each genotype, **P < 0.005). d Cumulative distribution plot and bar graph (mean ± s.e.m.) showing the mEPSC amplitude in MSNs from WT and CK1δ OE mice. e Cumulative distribution plot of the mEPSC inter-event interval and bar graph of mEPSC frequency in WT and OE mice. T-test, n = 3 animals/10 cells for each genotype, *P < 0.05. f Representative mEPSC traces. Scale bar: 10 pA, 1 s. g Bar graph summary of mean 10–90% rise time and decay time of mEPSC. h Cumulative plot and bar graph (mean ± s.e.m.) of mIPSC amplitude in MSNs from WT and CK1δ OE mice. i Cumulative plot of the mIPSC inter-event interval and bar graph of mIPSC frequency in WT and CK1δ OE mice. T-test, n = 3 animals/10 cells for each genotype, **P < 0.005. j Representative mIPSC traces. Scale bar: 20 pA, 1 s. k Bar graph summary of mean 10–90% rise time and decay time of mIPSC
Measurement of striatal volumes (using Golgi-stained sections) clearly showed that CK1δ OE mice have smaller striata than control littermates (Fig. 2c and Supplementary Figure S2c) (F1,10 = 10.72; P = 0.008) while the striatal neuronal densities between the two genotypes were not altered (data not shown). This finding matches patient brain imaging studies that find significant volume reductions of the regional caudate in naive patients, and attenuated abnormalities in the treated patients [2, 41].
To evaluate the role of CK1δ in neurotransmission, we investigated excitatory and inhibitory transmission in the MSNs of 5-week-old CK1δ OE mice and control littermates by recording mEPSCs and mIPSCs, respectively (Fig. 2d–k). The amplitude of mEPSCs was not altered in the CK1δ OE mice compared to control mice (Fig. 2d), indicating no change on the surface expression of glutamate receptors. But the mEPSC frequency was significantly increased in the CK1δ OE mice (Fig. 2e) (n = 3 mice per genotype, WT: 1.51 ± 0.29 Hz, n = 10 cells; OE: 2.61 ± 0.44 Hz, n = 10 cells, P = 0.020), suggesting an increase in the release probability of glutamate onto MSNs. Furthermore, for the inhibitory neurotransmission, the mIPSC amplitude was not altered in CK1δ OE mice (Fig. 2h), indicating unaltered surface level of GABA receptors. The mIPSC frequency was significantly decreased in the CK1δ OE mice compared to control mice (Fig. 2i) (n = 3 mice per genotype, WT: 2.5 ± 0.3 Hz, n = 10 cells; OE: 1.1 ± 0.1 Hz, n = 11 cells, P = 0.002), suggesting a decrease in the release probability of GABA onto MSNs. The kinetics of mEPSCs and mIPSCs were also analyzed and no change between CK1δ OE mice and control littermates was found (Fig. 2g, k). Our data suggest that glutamatergic and GABAergic synapses were not being redistributed. Taken together, these results suggest that CK1δ overexpression impairs the excitatory and inhibitory transmission in the striatal MSNs.
CK1δ OE mice exhibit altered synaptic ultrastructure in the striatum
Considering the structural modifications described above, we next investigated ultra-structurally the synapses of the mice using transmission electron microscopy. Micrographs of the striatum allowed us to visualize each of the four major synaptic types in both genotypes: (1) asymmetric synapses with predominately thick postsynaptic densities (PSDs) (Fig. 3a, b); (2) symmetric synapses containing thin PSDs (Fig. 3a, c); (3) perforated asymmetric synapses characterized as dashed PSD on a single postsynaptic spine (Fig. 3a, d); and (4) multi-contact synapses characterized as one single presynaptic terminal forming two or more postsynaptic elements with two or more dendritic spines (Fig. 3a, e). Asymmetric synapses predominately represent over 60–70% of the total synapses for both mouse genotypes (Fig. 3f). In contrast, the percentage of symmetric synapses are significantly different between the two genotypes. Indeed, symmetric synapses represent less than 20% in CK1δ OE mice while they represent almost 30% in control littermates (Fig. 3g) (F 1, 111 = 11.89; P = 0.0008).
Ultrastructural analysis of synapses in the striatum of CK1δ OE mice. a Representative TEM micrographs from WT and CK1δ OE mice, highlighting four kinds of synapses (see red boxes; b–e). b Asymmetric synapse (thick PSDs); c symmetric synapse (thin PSDs); d perforated synapse with dashed PSDs on the postsynaptic membrane in a single dendritic spine; and e multiple contact synapse (one axonal terminal forms two or more synapses with more than one dendritic spine). f–j Bar graphs showing the percentage of each type of synapse relative to the total number of synapses for each genotype (56 sections were analyzed corresponding to three mice for each genotype). f Asymmetric; g symmetric; h perforated; i multiple contacts; j total number of synapses. One-way ANOVA *P < 0.05, ***P < 0.001. Bar in a = 500 nm. b–e Red empty arrow heads and red arrows indicate presynaptic and postsynaptic compartments, respectively
Interestingly, perforated PSDs appear with a significantly higher frequency in the striatum of CK1δ OE mice (16.5% of total synapses) compared to control littermates (3%) (Fig. 3h) (F1, 111 = 12.686; P = 0.0007). Although CK1δ OE mice seem to harbor more multiple contact synapses, no significant difference was found between the two genotypes (Fig. 3i). In addition, the analysis of 56 electron micrographs (captured at ×10,000 magnification) from three mice for each genotype showed a significant decrease in the total number of synapses in the CK1δ OE compared to control littermates (229 versus 326; F1,111 = 15.478; P = 0.0001) (Fig. 3j).
The reduced number of inhibitory synapses in the striatum as observed here is consistent with the significant decrease of mIPSC frequency found in the CK1δ OE mice compared to control mice (Fig. 2i).
Genome-wide effects of CK1δ overexpression (OE) on gene expression
To determine what brain region to focus on for RNA-sequencing experiments, preliminary gene expression profiling was performed by microarray analysis and quantitative real-time PCR (qPCR). The microarrays results were analyzed using normalized expression values from Affymetrix Mouse Genome 430 2.0 arrays. Genes significantly altered (≥2-fold, OE versus WT) were determined using the Welch t-test and Benjamin Hochberg procedure (P < 0.05). Comparison of gene expression in the adult revealed that the striatum was the most significantly affected brain region with 160 genes significantly altered (77 up-regulated and 83 down-regulated) (Fig. 4a, Supplementary Table S1). Remarkably, a number of genes involved in the regulation of psychogenesis were up-regulated (e.g. Grm2, Adra2a, Htr1a, Gng2, Gng4, Geft, and Sox5) or down-regulated (e.g. Drd2, Adora2a, Adra2c, Gnb5, Gng7, and Traip). Their regulatory functions were positioned in the networks by IPA based on significance (P < 0.05), such as network relevant to ‘behavior’, and ‘nervous system development and function’ (Fig. 4b, left), and relevant to neurological diseases, psychological disorder and behavior performance (Fig. 4b, right). Only 31 genes (14 up-regulated and 17 down-regulated) in the fronto-cortex of CK1δ OE mice were found to be altered (Fig. 4a, Supplementary Table S2).
CK1δ OE altered gene expression in the striatum. a Bar chart shows the numbers of altered genes in the striatum and the frontal cortex in CK1δ OE mice, obtained using an Affymetrix genomic array, n = 3, P < 0.05 and fold changes >2. b Gene networks: (left) relevant to ‘behavior’, and ‘nervous system development and function’; (right) relevant to neurological diseases, psychological disorder, and behavior performance. Solid line presents gene interaction resulted from the Ingenuity Pathway Analysis (IPA). c Top canonical pathways (IPA) in which the genes are most altered after RNA sequencing. Gene fold changes ≥2 in the striatum are presented as –log 10 (P-value). The orange line indicates the threshold of P = 0.05, showing 15 out of 16 canonical pathways involved directly in CNS relevant functions. d Heatmap representation of 92 ADHD-candidate genes expression levels after RNAseq analysis shows altered expression of ADHD-risk genes in the striatum of OE versus WT mice. Red color: up-regulation; blue: down-regulation
The genes showed a differential expression in CK1δ OE mice, and those identified from the canonical pathway and network analysis were further validated by qPCR. Seventy-one genes showing altered expression, plus 45 genes directly related to these, and two controls (CK1δ and GAPDH) were tested (list of primers/genes Supplementary Table S3). Briefly, CK1δ was the most affected gene based on qPCR, in agreement with previous results [30]. Among all other CK1 isoforms only CK1γ3 was slightly down-regulated (Supplementary Figure S3a). Five isoforms of protein phosphatase 2c known to counteract with CK1 activity were confirmed to be up-regulated (Ppm1e, Ppm1h, ilkap, Ppm1l, PpmIm) (Supplementary Figure S3a). The isoforms of regulator of G-protein signaling (Rgs2, Rgs4, Rgs7, and Rgs9) and the enzymes GTPase-Rasd2 were significantly decreased (Supplementary Figure S3b). Dopamine receptor-mediated downstream signaling genes, CamKIIβ and Darpp-32, were down-regulated (Supplemetary Figure S3b). Both dopamine D2 receptor isoforms (Drd2s, presynaptic short isoform; Drd2l, postsynaptic long isoform) and Penk were down-regulated (Supplementary Figure S3c). The expression of Adra2a, Grm2, and Drd3 was up-regulated (Supplementary Figure S3c). The gene expression of several ion channels was also altered. Kcnc4, encoding Shaw-related subfamily member 4 of potassium voltage-gated channel, was significantly higher; Asic4, encoding a member of the acid-sensing ion channel family, were lower in the striatum and Scn4b, encoding voltage-gated sodium channel type IV β subunit (Supplementary Figure S3d). Calb1, St8sia3, and Fst were down-regulated while Calb2 and Vsnl1 were up-regulated (Supplementary Figure S3d). Furthermore, two genes encoding presynaptic proteins localized in the active zone, Syt13 and Doc2b, were also affected in CK1δ OE mice (Supplementary Figure S3e).
Based on these results, more extensive studies focusing on the striatum were carried out using deep sequencing (Illumina). Total mRNAs from the striata (four per genotype) were processed and submitted to high-throughput sequencing. The reads obtained were mapped and analyzed using HTSeq version 0.5.4p1 [42]. The principal component analysis (PCA) and unsupervised hierarchical clustering were performed to evaluate the relationship structure within-group and between-group. The values of FPKM (Fragments Per Kilobase of transcript per Million mapped reads) were transformed using log2(x + 1), and Benjamini-Hochberg adjusted P-values were calculated.
RNA sequencing data from striatal tissues showed that the expression of the genes involved in neuronal development processes are highly altered (Supplementary Table S4-S5). Importantly, dopamine signaling related genes such as D2 receptor, pre-pro-enkephalin, Penk, and Darpp-32 expression were significantly altered in CK1δ OE mice, confirming the results obtained by gene array analysis.
Canonical pathway analysis of genes for which the expression in the striatum was significantly altered(P < 0.05) was performed using the Ingenuity software. Remarkably, out of the top 16 canonical pathways that are most significantly altered (highest – log10 P-values), 15 are involved in CNS functions, such as cAMP-mediated signaling, Gαi signaling, 3G-protein coupled receptor and the dopamine-DARPP-32 pathway among others (Fig. 4c).
Quantitative genetic studies performed on ADHD patients [43, 44] have suggested a strong genetic component for this disorder. Collectively, at least 92 genes have been linked to ADHD risk in human through GWAS analysis (genome-wide association study) (Supplementary Table S5)[45,46,47,48,49,50]. We then explored a possible relationship between CK1δ OE-induced changes of gene expression and the 92 human genes genetically associated with ADHD. FPKM averages for each genotype corresponding to the 92 genes obtained by RNA sequencing from the striatum were plotted as a heatmap (Fig. 4d). Dramatic changes in the expression of most of these ADHD-candidate genes were observed in CK1δ OE mice compare to control mice (Fig. 4d; 4 left lanes compared to four right lanes, respectively). There is also a fairly good homogeneity of the results within genotype groups.
Gene expression profiling of Drd1a versus Drd2 MSNs, in CK1δ OE and WT mice
RNA-sequencing results indicate that dopaminergic signaling is perturbed and further reinforce the importance of dopamine in the mechanisms underlying ADHD. We next investigated the possibility that Drd1a and Drd2 MSNs might be affected differentially in CK1δ OE mice. Due to the high cellular heterogeneity and the intermixed nature of neuronal cell types in general, including for the striatum, Drd1a and Dr2d MSNs are highly intermixed and indistinguishable. Therefore, the molecular profiling of specific cell populations in vivo using bacTRAP [31, 51] was conducted. We engineered bacTRAP transgenic mice that enable cell type-specific molecular profiling of Drd1a and Drd2 neurons in the striatum of CK1δ OE versus WT mice by crossing CK1δ OE mice (tetO-CK1δ/Camk2α-tTA) with CP73 (Drd1a/EGFP-L10a) and CP101 (Drdr2/EGFP-L10a) mice. RNA profiling was performed on Drd1a and Drd2 neuronal populations from CK1δ OE and WT mice.
Two or three pairs of striata were pooled and three pools for each genotype were analyzed (7- 9 mice were used per genotype). Samples were processed and analyzed by deep sequencing (Illumina)(Supplementary Tables S6–S9). Top PCA resulted in a quadrant figure based on variance stabilizing transformed read counts of all genes and showed clear genotype and cell type separations (Fig. 5a). Heatmap representation of significantly altered gene expression between the four groups clearly demonstrated homeostasis perturbation (Fig. 5b). Only genes with Benjamini–Hochberg-adjusted P-values are shown, and FPKM values are transformed by log2(x + 1), and each row is scaled using Z-scores. The expression of 86 genes was altered (62 up-regulated and 24 down-regulated) in both cell types as indicated by the Venn diagram corresponding to altered genes (fold change cutoff = 2) between genotypes and for the two cell types tested (Fig. 5c, Supplementary Tables S7 and S9). The molecular types corresponding to the genes altered are shown in Supplementary Figure S4. Furthermore, canonical pathway analysis revealed that the top three pathways in each of the two cell types are different. Remarkably, the expressions of 70% or more of the genes included in these pathways are altered (Fig. 5d), and a significant fraction of the almost 100 genes identified as being linked to ADHD in different GWAS studies are differentially expressed in Drd1a and Drd2 MSNs compared the total pool of genes (Fisher exact: P = 0.0144 and P = 0.00139, respectively) (Supplementary Table S10). These results were confirmed individually by quantitative RT-PCR and revealed that, among 92 ADHD risk genes, eight are affected in both cell types (Adra2a to Zmiz1), 20 are specifically altered in the Drd1a cells, and 12 specifically altered in the Drd2 cells (Fig. 5e). CK1δ (Csnk1d) expression, as expected, increased in both cell types.
Differential gene alteration in Drd1a and Drd2 MSNs of CK1δ OE mice. a Principal component analysis was used to cluster samples based on gene expression. Each sample was generated by pooling two or three striatal tissues; three samples per genotype were analyzed for a total of 7–9 mice per condition. As expected samples corresponding to a specific genotype and cell type are clustering with each other. Noticeably, a very clear separation was found between the samples corresponding to the two cell types. b Heatmap of significantly altered genes in two genotypes and two cell types. Only genes with Benjamini–Hochberg-adjusted P-values are shown. FPKM values are transformed by log2(x + 1). Each row is scaled using Z-scores. c Venn diagram of altered genes (fold change cutoff = 2, and Benjamini–Hochberg-adjusted P-values <0.05) in the two cell types (red: up-regulated; green: down-regulated). d Top three canonical pathways in which the genes are altered in the Drd1a (left) and in the Drd2 (right) medium spiny neurons. e Genes identified by next-generation sequencing and corresponding to GWAS ADHD candidates have been validated by qPCR and then investigated for cell specificity using RNAseq data. Results corresponding to genes significantly altered in one or both cell types are shown (green: Drd1a and blue: Drd2)
Discussion
Attention refers to a process during which the central nervous system organizes sensory input and generates a coordinated behavioral response. Briefly, in the 3CSRT, food-deprived mice are required to pay attention to programmed visual stimuli and respond by correctly nose-poking in order to receive a food reward. This goal directed task requires visual sensory input processing and coordination of executive limbic motor neurons. The performance of CK1δ OE mice, particularly entering the food magazine more frequently during testing, demonstrated poor attention and low inhibition, underlying a combination of cognitive and sensory-processing deficits. These findings correlate well with some studies carried out on ADHD children, in which brain activation was measured through fMRI while performing behavioral tasks related to visual selective attention and response inhibition. Results showed that ADHD subjects made more errors compared to controls, and control subjects exhibited significantly greater intensity of activation in superior parietal and frontostriatal regions [52]. Regarding impulsivity, while we observed a trend in the effect of genotype on premature responses (P = 0.12), with OE mice exhibiting more premature responses compared to WT, it did not reach significance. This result might be partially due to the groups being underpowered as a result of very few OE mice (3 out of 10 mice) achieving criterion at all time points and completing the test.
d-Amphetamine, a psychostimulant drug reversing dopamine transporter action and leading to a release of large amounts of dopamine, has been typically used at low dose for the treatment of ADHD. While such treatment leads to addiction in non-ADHD subjects, it does not induce addiction in ADHD patients. Along those lines, brain imaging studies on ADHD patients revealed that ADHD brains show abnormal functional connectivity due to impaired interactions between control and reward pathways [12]. As we have shown previously, d-amphetamine effectively normalizes hyperactivity in the CK1δ OE mice and stimulates the control littermates [30]. For this reason, we chose d-amphetamine as the reinforcement in our place preference reward test and demonstrated that CK1δ OE mice display a lower association between contextual learning and reward, possibly lower motivation in drug searching, underpinning a disruption of the dopamine reward pathway.
Given the convergence of data in ADHD research, frontal cortex and striatum have become the prime targets for ADHD studies [53,54,55,56]. In CK1δ OE mice, shorter and less dense pyramidal neurons in the cingulate cortex may be responsible for the occurrence of inattentive behavior and poor nesting skills. Correspondingly, smaller striatal volume may further contribute to the disrupted neuro-circuitry leading to the impaired behavioral performances in CK1δ OE mice.
The pathology of ADHD has been linked to an abnormality of dopamine transmission. It is unknown if synaptic ultrastructural differences exist in ADHD brains. However, we describe here, in our mouse model, clear differences in the proportion of the four synaptic types, suggesting that some of those differences might contribute to the ADHD-like phenotypes. Asymmetric synapses are defined as excitatory synapses and symmetric synapses as inhibitory synapses [57,58,59]. Interestingly, in our mouse model we observed no difference in the amplitude, but an alteration in the frequency of excitatory and inhibitory synapses. The decrease in the inhibitory transmission and the increase in the excitatory transmission is compatible with the hyperactivity or lower motor inhibition observed. Furthermore, the frequent occurrences of aberrant perforated PSDs might reinforce abnormal neurotransmission [60,61,62,63]. It is unclear if perforated PSDs represent an intermediary stage of a spine, but based on the numbers of synapses observed in this study, the inverse correlation between higher number of perforated PSDs and lower number of total synapses would rather indicate that perforated PSDs correspond to a stable stage.
Despite ADHD’s association with dysfunction of dopaminergic cortico-subcortical networks related to executive functions and behavioral regulation, we cannot exclude that multiple events co-exist, either temporally or spatially, and they also could lead to the phenotypes described.
Perturbed homeostasis of gene transcriptome in the brain region of CK1δ OE mice underscores the fact that CK1δ is one of the important players in the occurrence of ADHD symptom. This could also explain the heterogeneity of symptoms observed in patients, and multi-genetic events occurring for ADHD onset and maintenance [45].
In conclusion, in the present study, we demonstrate that CK1δ OE mice exhibit a clear attention deficit and exhibit a disrupted dopamine reward pathway, highly reminiscent of ADHD. Using cell type-specific transcriptomic we demonstrated that the transcriptional homeostasis affects neurons directly, and that Drd1a and Drd2 MSNs are both affected and affected differently. All together, the neuronal abnormalities in the frontostriatal circuitry, the distorted striatal structures, the differences in synaptic connections, and the disrupted transcriptional homeostasis may underlie the mechanistic steps leading to ADHD pathophysiology. A schematic summary of the results is presented (Fig. 6). It will be interesting in future studies to investigate the role of CK1δ in developmental processes relevant to ADHD.
Summary of the phenotypes, differences, and alterations observed in CK1δ OE mice overexpressing CK1δ since conception. The top panels indicate the level of overexpression of CK1δ by in situ hybridization. The yellow lightning symbols indicate CK1 acute kinase activity. The blue arrows materialize the direct consequences of long-term CK1 overexpression, including developmental modifications. The doted blue arrows indicate possible secondary consequences. Yellow lightning symbols indicate acute CK1 activity
References
Castellanos FX, Proal E. Large-scale brain systems in ADHD: beyond the prefrontal-striatal model. Trends Cogn Sci. 2012;16:17–26.
Perou R, Bitsko RH, Blumberg SJ, Pastor P, Ghandour RM, Gfroerer JC, et al. Mental health surveillance among children—United States, 2005-2011. MMWR Suppl. 2013;62:1–35.
Advokat C. What are the cognitive effects of stimulant medications? Emphasis on adults with attention-deficit/hyperactivity disorder (ADHD). Neurosci Biobehav Rev. 2010;34:1256–66.
Wahlstrom D, Collins P, White T, Luciana M. Developmental changes in dopamine neurotransmission in adolescence: behavioral implications and issues in assessment. Brain Cogn. 2010;72:146–59.
Levy F, Hay DA, McStephen M, Wood C, Waldman I. Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. J Am Acad Child Adolesc Psychiatry. 1997;36:737–44.
Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9.
Zhao H, Nyholt DR. Gene-based analyses reveal novel genetic overlap and allelic heterogeneity across five major psychiatric disorders. Hum Genet. 2017;136:263–74.
Herrmann MJ, Biehl SC, Jacob C, Deckert J. Neurobiological and psychophysiological correlates of emotional dysregulation in ADHD patients. Atten Defic Hyperact Disord. 2010;2:233–9.
Rubia K. “Cool” inferior frontostriatal dysfunction in attention-deficit/hyperactivity disorder versus “hot” ventromedial orbitofrontal-limbic dysfunction in conduct disorder: a review. Biol Psychiatry. 2011;69:e69–87.
Rubia K, Halari R, Cubillo A, Smith AB, Mohammad AM, Brammer M, et al. Methylphenidate normalizes fronto-striatal underactivation during interference inhibition in medication-naive boys with attention-deficit hyperactivity disorder. Neuropsychopharmacology. 2011;36:1575–86.
Xia S, Foxe JJ, Sroubek AE, Branch C, Li X. Topological organization of the “small-world” visual attention network in children with attention deficit/hyperactivity disorder (ADHD). Front Hum Neurosci. 2014;8:162.
Tomasi D, Volkow ND. Abnormal functional connectivity in children with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2012;71:443–50.
Ernst M, Zametkin AJ, Matochik JA, Jons PH, Cohen RM. DOPA decarboxylase activity in attention deficit hyperactivity disorder adults. A [fluorine-18]fluorodopa positron emission tomographic study. J Neurosci. 1998;18:5901–7.
Ernst M, Zametkin AJ, Matochik JA, Pascualvaca D, Jons PH, Cohen RM. High midbrain [18F]DOPA accumulation in children with attention deficit hyperactivity disorder. Am J Psychiatry. 1999;156:1209–15.
Volkow ND, Wang GJ, Newcorn J, Fowler JS, Telang F, Solanto MV, et al. Brain dopamine transporter levels in treatment and drug naive adults with ADHD. Neuroimage. 2007;34:1182–90.
Volkow ND, Wang GJ, Kollins SH, Wigal TL, Newcorn JH, Telang F, et al. Evaluating dopamine reward pathway in ADHD: clinical implications. JAMA. 2009;302:1084–91.
Volkow ND, Wang GJ, Newcorn JH, Kollins SH, Wigal TL, Telang F, et al. Motivation deficit in ADHD is associated with dysfunction of the dopamine reward pathway. Mol Psychiatry. 2011;16:1147–54.
Morris G, Schmidt R, Bergman H. Striatal action-learning based on dopamine concentration. Exp Brain Res. 2010;200:307–17.
Tripp G, Wickens JR. Research review: dopamine transfer deficit: a neurobiological theory of altered reinforcement mechanisms in ADHD. J Child Psychol Psychiatry. 2008;49:691–704.
Luman M, Tripp G, Scheres A. Identifying the neurobiology of altered reinforcement sensitivity in ADHD: a review and research agenda. Neurosci Biobehav Rev. 2010;34:744–54.
Mattingly GW, Wilson J, Rostain AL. A clinician’s guide to ADHD treatment options. Postgrad Med. 2017;129:657–66.
Faraone SV, Biederman J, Wozniak J. Examining the comorbidity between attention deficit hyperactivity disorder and bipolar I disorder: a meta-analysis of family genetic studies. Am J Psychiatry. 2012;169:1256–66.
Leitner Y. The co-occurrence of autism and attention deficit hyperactivity disorder in children—what do we know? Front Hum Neurosci. 2014;8:268.
Desdouits F, Siciliano JC, Greengard P, Girault JA. Dopamine- and cAMP-regulated phosphoprotein DARPP-32: phosphorylation of Ser-137 by casein kinase I inhibits dephosphorylation of Thr-34 by calcineurin. Proc Natl Acad Sci USA. 1995;92:2682–5.
Gainetdinov RR, Wetsel WC, Jones SR, Levin ED, Jaber M, Caron MG. Role of serotonin in the paradoxical calming effect of psychostimulants on hyperactivity. Science. 1999;283:397–401.
Chergui K, Svenningsson P, Greengard P. Physiological role for casein kinase 1 in glutamatergic synaptic transmission. J Neurosci. 2005;25:6601–9.
Brennan KC, Bates EA, Shapiro RE, Zyuzin J, Hallows WC, Huang Y, et al. Casein kinase idelta mutations in familial migraine and advanced sleep phase. Sci Transl Med. 2013;5:1–11. 183ra56
Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–92.
Flajolet M, He G, Heiman M, Lin A, Nairn AC, Greengard P. Regulation of Alzheimer’s disease amyloid-beta formation by casein kinase I. Proc Natl Acad Sci USA. 2007;104:4159–64.
Zhou M, Rebholz H, Brocia C, Warner-Schmidt JL, Fienberg AA, Nairn AC, et al. Forebrain overexpression of CK1delta leads to down-regulation of dopamine receptors and altered locomotor activity reminiscent of ADHD. Proc Natl Acad Sci USA. 2010;107:4401–6.
Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–48.
Humby T, Laird FM, Davies W, Wilkinson LS. Visuospatial attentional functioning in mice: interactions between cholinergic manipulations and genotype. Eur J Neurosci. 1999;11:2813–23.
Krueger DD, Howell JL, Hebert BF, Olausson P, Taylor JR, Nairn AC. Assessment of cognitive function in the heterozygous reeler mouse. Psychopharmacol (Berl). 2006;189:95–104.
Cunningham CL, Gremel CM, Groblewski PA. Drug-induced conditioned place preference and aversion in mice. Nat Protoc. 2006;1:1662–70.
Bardo MT, Bevins RA. Conditioned place preference: what does it add to our preclinical understanding of drug reward? Psychopharmacol (Berl). 2000;153:31–43.
Bardo MT, Rowlett JK, Harris MJ. Conditioned place preference using opiate and stimulant drugs: a meta-analysis. Neurosci Biobehav Rev. 1995;19:39–51.
Feng HJ, Mathews GC, Kao C, Macdonald RL. Alterations of GABA A-receptor function and allosteric modulation during development of status epilepticus. J Neurophysiol. 2008;99:1285–93.
Lee JB, Wei J, Liu W, Cheng J, Feng J, Yan Z. Histone deacetylase 6 gates the synaptic action of acute stress in prefrontal cortex. J Physiol. 2012;590:1535–46.
Teicher MH, Anderson CM, Polcari A, Glod CA, Maas LC, Renshaw PF. Functional deficits in basal ganglia of children with attention-deficit/hyperactivity disorder shown with functional magnetic resonance imaging relaxometry. Nat Med. 2000;6:470–3.
Liotti M, Pliszka SR, Perez R, Kothmann D, Woldorff MG. Abnormal brain activity related to performance monitoring and error detection in children with ADHD. Cortex. 2005;41:377–88.
Rogers JH. Calretinin: a gene for a novel calcium-binding protein expressed principally in neurons. J Cell Biol. 1987;105:1343–53.
Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Thapar A, Martin J, Mick E, Arias Vasquez A, Langley K, Scherer SW, et al. Psychiatric gene discoveries shape evidence on ADHD’s biology. Mol Psychiatry. 2016;21:1202–7.
Turic D, Langley K, Williams H, Norton N, Williams NM, Moskvina V, et al. A family based study implicates solute carrier family 1-member 3 (SLC1A3) gene in attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1461–6.
Hawi Z, Cummins TD, Tong J, Johnson B, Lau R, Samarrai W, et al. The molecular genetic architecture of attention deficit hyperactivity disorder. Mol Psychiatry. 2015;20:289–97.
Yang L, Chang S, Lu Q, Zhang Y, Wu Z, Sun X, et al. A new locus regulating MICALL2 expression was identified for association with executive inhibition in children with attention deficit hyperactivity disorder. Mol Psychiatry. 2017;23:1014–20.
Bonvicini C, Faraone SV, Scassellati C. Attention-deficit hyperactivity disorder in adults: a systematic review and meta-analysis of genetic, pharmacogenetic and biochemical studies. Mol Psychiatry. 2016;21:872–84.
Hinney A, Scherag A, Jarick I, Albayrak O, Putter C, Pechlivanis S, et al. Genome-wide association study in German patients with attention deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:888–97.
Mastronardi CA, Pillai E, Pineda DA, Martinez AF, Lopera F, Velez JI, et al. Linkage and association analysis of ADHD endophenotypes in extended and multigenerational pedigrees from a genetic isolate. Mol Psychiatry. 2016;21:1434–40.
van der Voet M, Harich B, Franke B, Schenck A. ADHD-associated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol Psychiatry. 2016;21:565–73.
Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–62.
Booth JR, Burman DD, Meyer JR, Lei Z, Trommer BL, Davenport ND, et al. Larger deficits in brain networks for response inhibition than for visual selective attention in attention deficit hyperactivity disorder (ADHD). J Child Psychol Psychiatry. 2005;46:94–111.
Seidman LJ, Valera EM, Makris N, Monuteaux MC, Boriel DL, Kelkar K, et al. Dorsolateral prefrontal and anterior cingulate cortex volumetric abnormalities in adults with attention-deficit/hyperactivity disorder identified by magnetic resonance imaging. Biol Psychiatry. 2006;60:1071–80.
Spencer TJ, Biederman J, Madras BK, Faraone SV, Dougherty DD, Bonab AA, et al. In vivo neuroreceptor imaging in attention-deficit/hyperactivity disorder: a focus on the dopamine transporter. Biol Psychiatry. 2005;57:1293–300.
Castellanos FX, Tannock R. Neuroscience of attention-deficit/hyperactivity disorder: the search for endophenotypes. Nat Rev Neurosci. 2002;3:617–28.
Bush G, Valera EM, Seidman LJ. Functional neuroimaging of attention-deficit/hyperactivity disorder: a review and suggested future directions. Biol Psychiatry. 2005;57:1273–84.
Beaulieu C, Colonnier M. A laminar analysis of the number of round-asymmetrical and flat-symmetrical synapses on spines, dendritic trunks, and cell bodies in area 17 of the cat. J Comp Neurol. 1985;231:180–9.
Yung KK, Bolam JP, Smith AD, Hersch SM, Ciliax BJ, Levey AI. Immunocytochemical localization of D1 and D2 dopamine receptors in the basal ganglia of the rat: light and electron microscopy. Neuroscience. 1995;65:709–30.
Delle Donne KT, Sesack SR, Pickel VM. Ultrastructural immunocytochemical localization of the dopamine D2 receptor within GABAergic neurons of the rat striatum. Brain Res. 1997;746:239–55.
Ganeshina O, Berry RW, Petralia RS, Nicholson DA, Geinisman Y. Differences in the expression of AMPA and NMDA receptors between axospinous perforated and nonperforated synapses are related to the configuration and size of postsynaptic densities. J Comp Neurol. 2004;468:86–95.
Geinisman Y. Perforated axospinous synapses with multiple, completely partitioned transmission zones: probable structural intermediates in synaptic plasticity. Hippocampus. 1993;3:417–33.
Yankova M, Hart SA, Woolley CS. Estrogen increases synaptic connectivity between single presynaptic inputs and multiple postsynaptic CA1 pyramidal cells: a serial electron-microscopic study. Proc Natl Acad Sci USA. 2001;98:3525–30.
Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, et al. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462:1065–9.
Acknowledgements
We would like to thank Drs. Jack Zhang and Wei Wang for their help analyzing deep sequencing data and 3CSRT data. We are grateful to Drs. Angus Nairn and Jean-Pierre Roussarie for helpful discussions. We thank Mallory Kerner, Julia Fram, and Randall Tassone for technical assistance. We would like to acknowledge the animal care and veterinary staff at RU for their excellent animal support, and especially Janelle Monnas, Craig Hunter, and Alejandra Gonzalez. This work was supported in part by National Institutes of Health Grants MH090963, DA010044, AG047781, by The Army Medical Research and Materiel Command (W81XWH-10-1-0691 to M.F.), and by the Black family foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhou, M., Gresack, J., Cheng, J. et al. CK1δ over-expressing mice display ADHD-like behaviors, frontostriatal neuronal abnormalities and altered expressions of ADHD-candidate genes. Mol Psychiatry 25, 3322–3336 (2020). https://doi.org/10.1038/s41380-018-0233-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-018-0233-z