Abstract
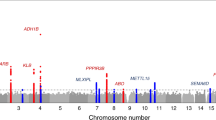
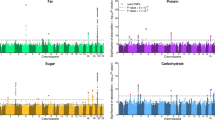
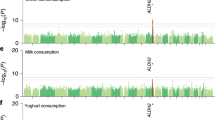
Macronutrient intake, the proportion of calories consumed from carbohydrate, fat, and protein, is an important risk factor for metabolic diseases with significant familial aggregation. Previous studies have identified two genetic loci for macronutrient intake, but incomplete coverage of genetic variation and modest sample sizes have hindered the discovery of additional loci. Here, we expanded the genetic landscape of macronutrient intake, identifying 12 suggestively significant loci (P < 1 × 10−6) associated with intake of any macronutrient in 91,114 European ancestry participants. Four loci replicated and reached genome-wide significance in a combined meta-analysis including 123,659 European descent participants, unraveling two novel loci; a common variant in RARB locus for carbohydrate intake and a rare variant in DRAM1 locus for protein intake, and corroborating earlier FGF21 and FTO findings. In additional analysis of 144,770 participants from the UK Biobank, all identified associations from the two-stage analysis were confirmed except for DRAM1. Identified loci might have implications in brain and adipose tissue biology and have clinical impact in obesity-related phenotypes. Our findings provide new insight into biological functions related to macronutrient intake.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ezzati M, Riboli E. Behavioral and dietary risk factors for noncommunicable diseases. N Engl J Med. 2013;369:954–64.
U.S. Department of Health and Human Services and U.S. Department of Agriculture. 2015 – 2020 Dietary Guidelines for Americans. 8th edn. December 2015. https://health.gov/dietaryguidelines/2015/guidelines/.
The Eatwell Guide -GOV.UK. https://www.gov.uk/government/publications/the-eatwell-guide
Montagnese C, Santarpia L, Buonifacio M, Nardelli A, Caldara AR, Silvestri E, et al. European food-based dietary guidelines: a comparison and update. Nutrition. 2015;31:908–15.
Food based dietary guidelines in the WHO European Region Nutrition and Food Security Programme WHO Regional Office for Europe Scherfigsvej 8, 2100 Copenhagen Denmark; http://www.euro.who.int/__data/assets/pdf_file/0017/150083/E79832.pdf
de Castro JM. The control of food intake of free-living humans: putting the pieces back together. Physiol Behav. 2010;100:446–53.
Rankinen T, Bouchard C. Genetics of food intake and eating behavior phenotypes in humans. Annu Rev Nutr. 2006;26:413–34.
Teucher B, Skinner J, Skidmore PML, Cassidy A, Fairweather-Tait SJ, Hooper L, et al. Dietary patterns and heritability of food choice in a UK female twin cohort. Twin Res Hum Genet. 2007;10:734–48.
Chu AY, Workalemahu T, Paynter NP, Rose LM, Giulianini F, Tanaka T, et al. Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet. 2013;22:1895–902.
Søberg S, Sandholt CH, Jespersen NZ, Toft U, Madsen AL, von Holstein-Rathlou S, et al. FGF21 Is a sugar-induced hormone associated with sweet intake and preference in humans. Cell Metab. 2017;25:1045–53.e6.
Potthoff MJ. FGF21 and metabolic disease in 2016: a new frontier in FGF21 biology. Nat Rev Endocrinol. 2017;13:74–6.
Tanaka T, Ngwa JS, van Rooij FJA, Zillikens MC, Wojczynski MK, Frazier-Wood AC, et al. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97:1395–402.
Spencer CCA, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
de Vries PS, Sabater-Lleal M, Chasman DI, Trompet S, Ahluwalia TS, Teumer A, et al. Comparison of HapMap and 1000 genomes reference panels in a large-scale genome-wide association study. Yao Y-G, editor. PLoS ONE. 2017;12:e0167742.
Manolio TA, Weis BK, Cowie CC, Hoover RN, Hudson K, Kramer BS, et al. New models for large prospective studies: is there a better way? Am J Epidemiol. 2012;175:859–66.
Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34.
Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4.
Delaneau O, Marchini J, 1000 Genomes Project Consortium GA, 1000 Genomes Project Consortium P, Lunter G, Marchini JL, et al. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934.
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1.
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium N. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66.
Huang Y-F, Gulko B, Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 2017;49:618–24.
Kichaev G, Yang W-Y, Lindstrom S, Hormozdiari F, Eskin E, Price AL, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. Di Rienzo A, editor. PLoS Genet. 2014;10:e1004722.
Ardlie KG, Deluca DS, Segre AV, Sullivan TJ, Young TR, Gelfand ET, et al. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60.
Hon C-C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature. 2017;543:199–204.
Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–9.
T2D-GENES Consortium, GoT2D Consortium, DIAGRAM Consortium. 2017.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14.
Ford ES, Dietz WH. Trends in energy intake among adults in the United States: findings from NHANES. Am J Clin Nutr. 2013;97:848–53.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Abajobir AA, Abate KH, Abbafati C, Abbas KM, Abd-Allah F, Abdulle AM, et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1345–422.
Basu S, Yoffe P, Hills N, Lustig RH. The relationship of sugar to population-level diabetes prevalence: an econometric analysis of repeated cross-sectional data. PLoS ONE. 2013;8:e57873.
Siervo M, Montagnese C, Mathers JC, Soroka KR, Stephan BCM, Wells JCK. Sugar consumption and global prevalence of obesity and hypertension: an ecological analysis. Public Health Nutr. 2014;17:587–96.
Volkow ND, Wang G-J, Baler RD. Reward, dopamine and the control of food intake: implications for obesity. Trends Cogn Sci. 2011;15:37–46.
Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34.
Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun. 2015;6:5897.
Almgren P, Lindqvist A, Krus U, Hakaste L, Ottosson-Laakso E, Asplund O, et al. Genetic determinants of circulating GIP and GLP-1 concentrations. JCI insight. 2017;2:e93306.
Heitmann BL, Lissner L. Dietary underreporting by obese individuals--is it specific or non-specific? BMJ. 1995;311:986–9.
Qi Q, Kilpeläinen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, et al. FTO genetic variants, dietary intake, and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;23:6961–72.
Johnston BC, Kanters S, Bandayrel K, Wu P, Naji F, Siemieniuk Ra, et al. Comparison of weight loss among named diet programs in overweight and obese adults. JAMA. 2014;312:923–33.
Murray CJL, Atkinson C, Bhalla K, Birbeck G, Burstein R, Chou D, et al. The state of US health, 1990-2010: burden of diseases, injuries, and risk factors. JAMA. 2013;310:591–608.
Hu FB, Satija A, Manson JE. Curbing the diabetes pandemic: the need for global policy solutions. JAMA. 2015;313:2319–20.
Barnard ND, Willett WC, Ding EL. The misuse of meta-analysis in nutrition research. JAMA. 2017;318:1435.
Acknowledgements
A full list of acknowledgments appears in Supplementary Table 6. We acknowledge the essential role of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium for encouraging CHARGE studies to participate in this effort and for the contributions of CHARGE members to the analyses conducted for this research. Dr. Caren Smith is supported by NHLBI K08 HL112845.
Authors contributions
D.I.C., T.T., and A.Y.C. conceived and designed the study. A.E.J., M.G., C.P., G.V.D., R.N.L., M.K.W., S.M., J.S.N., M.K., T.S.A., N.P., D.K.H., C.B., T.H., M.O., A.C.F.-W., D.O.M.-K., L.P., C.E.P., P.S.V., T.V., O.L., S.K., L.M.R., T.L., J.H.Z., M.F.F., J.L., N.M.M., J.A.S., T.H., N.E., M.A.N., T.R., J.H., D.G.H., C.-A.S., A.M., R.L.-G., M.-C.V., C.A.W., F.J.A.R., J.S., I.P.K., F.D., P.M.R., M.K., D.S.S., C.L., W.Z., A.A., P.K., M.G., D.C.R., Q.Q., L.F., U.E., J.B., A.H., Z.P., V.M., N.J.W., S.L.R.K., O.P., A.J., J.E.C., M.C.Z., J.S.V., N.G.F., J.M.O., J.C.L., H.R., A.G.U., O.T.R., J.C.K.-d.J., J.D., J.I.R., K.W.N., R.A.S., M.A.P., M.P., L.A.C., S.T.T., T.I.A.S., V.K., Y.L., Y.S., L.Q., S.B., S.S.R., R.M., A.T., W.H.O., O.H.F., T.P., J.C.F., P.D., L.-P.L., contributed to data collection. J.M., H.S.D., D.I.C., T.T., and A.Y.C. performed quality control and meta-analyses. J.M., A.E.J., C.S., S.X.L. performed additional analyses including look-up in the NH.G.R.I.-E.B.I. G.W.A.S. Catalog, fine-mapping analyses, colocalization analyses for Expression Quantitative Trait loci and Human Atlas of long non-coding R.N.A., L.D. score regression analyses and Mendelian randomization. J.M., H.S.D., S.X.L., A.E.J., M.G., C.P., C.E.S., G.V.D., D.I.C., T.T., and A.Y.C. wrote the manuscript. All authors approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
MANs’ participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging, NIH, Bethesda, MD, USA, as a possible conflict of interest. Dr. Nalls also consults for Illumina Inc, the Michael J. Fox Foundation and University of California Healthcare among others. AYC is currently employed by Merck Research Laboratories.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Merino, J., Dashti, H.S., Li, S.X. et al. Genome-wide meta-analysis of macronutrient intake of 91,114 European ancestry participants from the cohorts for heart and aging research in genomic epidemiology consortium. Mol Psychiatry 24, 1920–1932 (2019). https://doi.org/10.1038/s41380-018-0079-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-018-0079-4
This article is cited by
-
Genetic correlation and Mendelian randomization analyses support causal relationships between dietary habits and age at menarche
Scientific Reports (2024)
-
Genetic predisposition to macronutrient preference and workplace food choices
Molecular Psychiatry (2023)
-
RNA methylation in vascular disease: a systematic review
Journal of Cardiothoracic Surgery (2022)
-
Cardiovascular and cancer risk factors analysis for 2001–2020 from the global research output and European newspapers
Scientometrics (2022)
-
A genome-wide association study on adherence to low-carbohydrate diets in Japanese
European Journal of Clinical Nutrition (2022)