Abstract
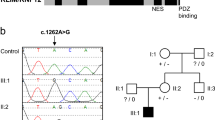
RLIM, also known as RNF12, is an X-linked E3 ubiquitin ligase acting as a negative regulator of LIM-domain containing transcription factors and participates in X-chromosome inactivation (XCI) in mice. We report the genetic and clinical findings of 84 individuals from nine unrelated families, eight of whom who have pathogenic variants in RLIM (RING finger LIM domain-interacting protein). A total of 40 affected males have X-linked intellectual disability (XLID) and variable behavioral anomalies with or without congenital malformations. In contrast, 44 heterozygous female carriers have normal cognition and behavior, but eight showed mild physical features. All RLIM variants identified are missense changes co-segregating with the phenotype and predicted to affect protein function. Eight of the nine altered amino acids are conserved and lie either within a domain essential for binding interacting proteins or in the C-terminal RING finger catalytic domain. In vitro experiments revealed that these amino acid changes in the RLIM RING finger impaired RLIM ubiquitin ligase activity. In vivo experiments in rlim mutant zebrafish showed that wild type RLIM rescued the zebrafish rlim phenotype, whereas the patient-specific missense RLIM variants failed to rescue the phenotype and thus represent likely severe loss-of-function mutations. In summary, we identified a spectrum of RLIM missense variants causing syndromic XLID and affecting the ubiquitin ligase activity of RLIM, suggesting that enzymatic activity of RLIM is required for normal development, cognition and behavior.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Retaux S, Bachy I. A short history of LIM domains (1993-2002): from protein interaction to degradation. Mol Neurobiol. 2002;26:269–81.
Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van IW, et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–90.
Hu H, Haas SA, Chelly J, Van Esch H, Raynaud M, de Brouwer AP, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry. 2016;21:133–48.
Tonne E, Holdhus R, Stansberg C, Stray-Pedersen A, Petersen K, Brunner HG, et al. Syndromic X-linked intellectual disability segregating with a missense variant in RLIM. Eur J Hum Genet. 2015;23:1652–6.
Rodriguez Criado G. [New X linked mental retardation syndrome]. An De Pediatr. 2012;76:184–191.
Gao R, Wang L, Cai H, Zhu J, Yu L. E3 ubiquitin ligase RLIM negatively regulates c-Myc transcriptional activity and restrains cell proliferation. PloS ONE. 2016;11:e0164086.
Ostendorff HP, Bossenz M, Mincheva A, Copeland NG, Gilbert DJ, Jenkins NA, et al. Functional characterization of the gene encoding RLIM, the corepressor of LIM homeodomain factors. Genomics. 2000;69:120–30.
Bach I, Rodriguez-Esteban C, Carriere C, Bhushan A, Krones A, Rose DW, et al. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat Genet. 1999;22:394–9.
Ostendorff HP, Peirano RI, Peters MA, Schluter A, Bossenz M, Scheffner M, et al. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103.
Hiratani I, Yamamoto N, Mochizuki T, Ohmori SY, Taira M. Selective degradation of excess Ldb1 by Rnf12/RLIM confers proper Ldb1 expression levels and Xlim-1/Ldb1 stoichiometry in Xenopus organizer functions. Development. 2003;130:4161–75.
Zhang L, Huang H, Zhou F, Schimmel J, Pardo CG, Zhang T, et al. RNF12 controls embryonic stem cell fate and morphogenesis in zebrafish embryos by targeting Smad7 for degradation. Mol Cell. 2012;46:650–61.
Shin J, Wallingford MC, Gallant J, Marcho C, Jiao B, Byron M, et al. RLIM is dispensable for X-chromosome inactivation in the mouse embryonic epiblast. Nature. 2014;511:86–89.
Ostendorff HP, Tursun B, Cornils K, Schluter A, Drung A, Gungor C, et al. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev Dyn. 2006;235:786–91.
Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, et al. RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011.
Barakat TS, Loos F, van Staveren S, Myronova E, Ghazvini M, Grootegoed JA, et al. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell. 2014;53:965–78.
Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, et al. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–81.
Gontan C, Jonkers I, Gribnau J. Long noncoding RNAs and X chromosome inactivation. Prog Mol Subcell Biol. 2011;51:43–64.
Her YR, Chung IK. Ubiquitin ligase RLIM modulates telomere length homeostasis through a proteolysis of TRF1. J Biol Chem. 2009;284:8557–66.
Chen X, Shen J, Li X, Wang X, Long M, Lin F, et al. Rlim, an E3 ubiquitin ligase, influences the stability of Stathmin protein in human osteosarcoma cells. Cell Signal. 2014;26:1532–8.
Gao K, Wang C, Jin X, Xiao J, Zhang E, Yang X, et al. RNF12 promotes p53-dependent cell growth suppression and apoptosis by targeting MDM2 for destruction. Cancer Lett. 2016;375:133–41.
Huang Y, Yang Y, Gao R, Yang X, Yan X, Wang C, et al. RLIM interacts with Smurf2 and promotes TGF-beta induced U2OS cell migration. Biochem Biophys Res Commun. 2011;414:181–5.
Johnsen SA, Gungor C, Prenzel T, Riethdorf S, Riethdorf L, Taniguchi-Ishigaki N, et al. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 2009;69:128–36.
Jiao B, Taniguchi-Ishigaki N, Gungor C, Peters MA, Chen YW, Riethdorf S, et al. Functional activity of RLIM/Rnf12 is regulated by phosphorylation-dependent nucleocytoplasmic shuttling. Mol Biol Cell. 2013;24:3085–96.
Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704.
Weese D, Emde AK, Rausch T, Doring A, Reinert K. RazerS--fast read mapping with sensitivity control. Genome Res. 2009;19:1646–54.
Emde AK, Schulz MH, Weese D, Sun R, Vingron M, Kalscheuer VM, et al. Detecting genomic indel variants with exact breakpoints in single- and paired-end sequencing data using SplazerS. Bioinformatics. 2012;28:619–27.
Kelly M, Williams R, Aojula A, O'Neill J, Trzinscka Z, Grover L, et al. Peptide aptamers: novel coatings for orthopaedic implants. Mater Sci Eng C Mater Biol Appl. 2015;54:84–93.
des Portes V, Beldjord C, Chelly J, Hamel B, Kremer H, Smits A, et al. X-linked nonspecific mental retardation (MRX) linkage studies in 25 unrelated families: the European XLMR consortium. Am J Med Genet. 1999;85:263–5.
Frints SG, Borghgraef M, Froyen G, Marynen P, Fryns JP. Clinical study and haplotype analysis in two brothers with Partington syndrome. Am J Med Genet. 2002;112:361–8.
Carrel L, Willard HF. An assay for X inactivation based on differential methylation at the fragile X locus, FMR1. Am J Med Genet. 1996;64:27–30.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–39.
Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 3rd ed., vol. 385. Eugene, OR: University of Oregon Press; 1995.
Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–9.
Donahoe PK, Longoni M, High FA. Polygenic causes of congenital diaphragmatic hernia produce common lung pathologies. Am J Pathol. 2016;186:2532–43.
Yu L, Sawle AD, Wynn J, Aspelund G, Stolar CJ, Arkovitz MS, et al. Increased burden of de novo predicted deleterious variants in complex congenital diaphragmatic hernia. Hum Mol Genet. 2015;24:4764–73.
Longoni M, High FA, Qi H, Joy MP, Hila R, Coletti CM, et al. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum Genet. 2017;136:679–91.
Longoni M, High FA, Russell MK, Kashani A, Tracy AA, Coletti CM, et al. Molecular pathogenesis of congenital diaphragmatic hernia revealed by exome sequencing, developmental data, and bioinformatics. Proc Natl Acad Sci USA. 2014;111:12450–5.
Mullegama SV, Klein SD, Mulatinho MV, Senaratne TN, Singh K, Center UCG, et al. De novo loss-of-function variants in STAG2 are associated with developmental delay, microcephaly, and congenital anomalies. Am J Med Genet A. 2017;173:1319–27.
Kumar R, Corbett MA, Van Bon BW, Gardner A, Woenig JA, Jolly LA, et al. Increased STAG2 dosage defines a novel cohesinopathy with intellectual disability and behavioral problems. Hum Mol Genet. 2015;24:7171–81.
Yingjun X, Wen T, Yujian L, Lingling X, Huimin H, Qun F, et al. Microduplication of chromosome Xq25 encompassing STAG2 gene in a boy with intellectual disability. Eur J Med Genet. 2015;58:116–21.
Bone KM, Chernos JE, Perrier R, Innes AM, Bernier FP, McLeod R, et al. Mosaic trisomy 1q: a recurring chromosome anomaly that is a diagnostic challenge and is associated with a Fryns-like phenotype. Prenat Diagn. 2017;37:602–10.
Rossetti R, Ferrari I, Bonomi M, Persani L. Genetics of primary ovarian insufficiency. Clin Genet. 2017;91:183–98.
Bach I. Releasing the break on X chromosome inactivation: Rnf12/RLIM targets REX1 for degradation. Cell Res. 2012;22:1524–6.
Upadhyay A, Joshi V, Amanullah A, Mishra R, Arora N, Prasad A, et al. E3 ubiquitin ligases neurobiological mechanisms: development to degeneration. Front Mol Neurosci. 2017;10:151.
Margolin DH, Kousi M, Chan YM, Lim ET, Schmahmann JD, Hadjivassiliou M, et al. Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N Engl J Med. 2013;368:1992–2003.
Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73.
Basel-Vanagaite L, Dallapiccola B, Ramirez-Solis R, Segref A, Thiele H, Edwards A, et al. Deficiency for the ubiquitin ligase UBE3B in a blepharophimosis-ptosis-intellectual-disability syndrome. Am J Hum Genet. 2012;91:998–1010.
Nascimento RM, Otto PA, de Brouwer AP, Vianna-Morgante AM. UBE2A, which encodes a ubiquitin-conjugating enzyme, is mutated in a novel X-linked mental retardation syndrome. Am J Hum Genet. 2006;79:549–55.
Budny B, Badura-Stronka M, Materna-Kiryluk A, Tzschach A, Raynaud M, Latos-Bielenska A, et al. Novel missense mutations in the ubiquitination-related gene UBE2A cause a recognizable X-linked mental retardation syndrome. Clin Genet. 2010;77:541–51.
Vandewalle J, Bauters M, Van Esch H, Belet S, Verbeeck J, Fieremans N, et al. The mitochondrial solute carrier SLC25A5 at Xq24 is a novel candidate gene for non-syndromic intellectual disability. Hum Genet. 2013;132:1177–85.
Haddad DM, Vilain S, Vos M, Esposito G, Matta S, Kalscheuer VM, et al. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol Cell. 2013;50:831–43.
Flex E, Ciolfi A, Caputo V, Fodale V, Leoni C, Melis D, et al. Loss of function of the E3 ubiquitin-protein ligase UBE3B causes Kaufman oculocerebrofacial syndrome. J Med Genet. 2013;50:493–9.
Isrie M, Kalscheuer VM, Holvoet M, Fieremans N, Van Esch H, Devriendt K. HUWE1 mutation explains phenotypic severity in a case of familial idiopathic intellectual disability. Eur J Med Genet. 2013;56:379–82.
Froyen G, Belet S, Martinez F, Santos-Reboucas CB, Declercq M, Verbeeck J, et al. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am J Hum Genet. 2012;91:252–64.
Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, Meldrum C, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–43.
So J, Suckow V, Kijas Z, Kalscheuer V, Moser B, Winter J, et al. Mild phenotypes in a series of patients with Opitz GBBB syndrome with MID1 mutations. Am J Med Genet A. 2005;132A:1–7.
Quaderi NA, Schweiger S, Gaudenz K, Franco B, Rugarli EI, Berger W, et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17:285–91.
Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, Ropers HH, et al. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet. 2001;29:287–94.
Schweiger S, Dorn S, Fuchs M, Kohler A, Matthes F, Muller EC, et al. The E3 ubiquitin ligase MID1 catalyzes ubiquitination and cleavage of Fu. J Biol Chem. 2014;289:31805–17.
Zou Y, Liu Q, Chen B, Zhang X, Guo C, Zhou H, et al. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am J Hum Genet. 2007;80:561–6.
Vulto-van Silfhout AT, Nakagawa T, Bahi-Buisson N, Haas SA, Hu H, Bienek M, et al. Variants in CUL4B are associated with cerebral malformations. Hum Mutat. 2015;36:106–17.
Badura-Stronka M, Jamsheer A, Materna-Kiryluk A, Sowinska A, Kiryluk K, Budny B, et al. A novel nonsense mutation in CUL4B gene in three brothers with X-linked mental retardation syndrome. Clin Genet. 2010;77:141–4.
Tarpey PS, Raymond FL, O'Meara S, Edkins S, Teague J, Butler A, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet. 2007;80:345–52.
Hwang CS, Sukalo M, Batygin O, Addor MC, Brunner H, Aytes AP, et al. Ubiquitin ligases of the N-end rule pathway: assessment of mutations in UBR1 that cause the Johanson-Blizzard syndrome. PLoS ONE. 2011;6:e24925.
Sukalo M, Fiedler A, Guzman C, Spranger S, Addor MC, McHeik JN, et al. Mutations in the human UBR1 gene and the associated phenotypic spectrum. Hum Mutat. 2014;35:521–31.
Zenker M, Mayerle J, Lerch MM, Tagariello A, Zerres K, Durie PR, et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat Genet. 2005;37:1345–50.
Zhang J, Gambin T, Yuan B, Szafranski P, Rosenfeld JA, Balwi MA, et al. Erratum to: Haploinsufficiency of the E3 ubiquitin-protein ligase gene TRIP12 causes intellectual disability with or without autism spectrum disorders, speech delay, and dysmorphic features. Hum Genet. 2017;136:1009–11.
Zhang J, Gambin T, Yuan B, Szafranski P, Rosenfeld JA, Balwi MA, et al. Haploinsufficiency of the E3 ubiquitin-protein ligase gene TRIP12 causes intellectual disability with or without autism spectrum disorders, speech delay, and dysmorphic features. Hum Genet. 2017;136:377–86.
Bramswig NC, Ludecke HJ, Pettersson M, Albrecht B, Bernier RA, Cremer K, et al. Identification of new TRIP12 variants and detailed clinical evaluation of individuals with non-syndromic intellectual disability with or without autism. Hum Genet. 2017;136:179–92.
Alqwaifly M, Bohlega S. Ataxia and hypogonadotropic hypogonadism with intrafamilial variability caused by RNF216 mutation. Neurol Int. 2016;8:6444.
Ganos C, Hersheson J, Adams M, Bhatia KP, Houlden H. Syndromic associations and RNF216 mutations. Park Relat Disord. 2015;21:1389–90.
Santens P, Van Damme T, Steyaert W, Willaert A, Sablonniere B, De Paepe A, et al. RNF216 mutations as a novel cause of autosomal recessive Huntington-like disorder. Neurology. 2015;84:1760–6.
Kramer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–20.
Son MY, Kwak JE, Kim YD, Cho YS. Proteomic and network analysis of proteins regulated by REX1 in human embryonic stem cells. Proteomics. 2015;15:2220–9.
Hill CS. Inhibiting the inhibitor: the role of RNF12 in TGF-beta superfamily signaling. Mol Cell. 2012;46:558–9.
Stricker S, Mundlos S. Mechanisms of digit formation: human malformation syndromes tell the story. Dev Dyn. 2011;240:990–1004.
Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1:87–105.
Emmerton-Coughlin HM, Martin KK, Chiu JS, Zhao L, Scott LA, Regnault TR, et al. BMP4 and LGL1 are down regulated in an ovine model of congenital diaphragmatic hernia. Front Surg. 2014;1:44.
Papangeli I, Scambler PJ. Tbx1 genetically interacts with the transforming growth factor-beta/bone morphogenetic protein inhibitor Smad7 during great vessel remodeling. Circ Res. 2013;112:90–102.
Mesbah K, Rana MS, Francou A, van Duijvenboden K, Papaioannou VE, Moorman AF, et al. Identification of a Tbx1/Tbx2/Tbx3 genetic pathway governing pharyngeal and arterial pole morphogenesis. Hum Mol Genet. 2012;21:1217–29.
Weider M, Wegner M. SoxE factors: transcriptional regulators of neural differentiation and nervous system development. Semin Cell Dev Biol. 2017;63:35–42.
Sha L, MacIntyre L, Machell JA, Kelly MP, Porteous DJ, Brandon NJ, et al. Transcriptional regulation of neurodevelopmental and metabolic pathways by NPAS3. Mol Psychiatry. 2012;17:267–79.
Navarro P, Moffat M, Mullin NP, Chambers I. The X-inactivation trans-activator Rnf12 is negatively regulated by pluripotency factors in embryonic stem cells. Human Genet. 2011;130:255–64.
Marie PJ, Debiais F, Hay E. Regulation of human cranial osteoblast phenotype by FGF-2, FGFR-2 and BMP-2 signaling. Histol Histopathol. 2002;17:877–85.
Ishibashi M, Saitsu H, Komada M, Shiota K. Signaling cascade coordinating growth of dorsal and ventral tissues of the vertebrate brain, with special reference to the involvement of Sonic Hedgehog signaling. Anat Sci Int. 2005;80:30–36.
Moen MJ, Adams HH, Brandsma JH, Dekkers DH, Akinci U, Karkampouna S, et al. An interaction network of mental disorder proteins in neural stem cells. Transl Psychiatry. 2017;7:e1082.
Yu L, Arbez N, Nucifora LG, Sell GL, Delisi LE, Ross CA, et al. A mutation in NPAS3 segregates with mental illness in a small family. Mol Psychiatry. 2014;19:7–8.
Nucifora LG, Wu YC, Lee BJ, Sha L, Margolis RL, Ross CA, et al. A mutation in NPAS3 that segregates with schizophrenia in a small family leads to protein aggregation. Mol Neuropsychiatry. 2016;2:133–44.
Pickard BS, Christoforou A, Thomson PA, Fawkes A, Evans KL, Morris SW, et al. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:874–84.
Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8.
Lavedan C, Licamele L, Volpi S, Hamilton J, Heaton C, Mack K, et al. Association of the NPAS3 gene and five other loci with response to the antipsychotic iloperidone identified in a whole genome association study. Mol Psychiatry. 2009;14:804–19.
Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, et al. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci USA. 2004;101:13648–53.
Zhou S, Degan S, Potts EN, Foster WM, Sunday ME. NPAS3 is a trachealess homolog critical for lung development and homeostasis. Proc Natl Acad Sci USA. 2009;106:11691–6.
Witteveen JS, Willemsen MH, Dombroski TC, van Bakel NH, Nillesen WM, van Hulten JA, et al. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat Genet. 2016;48:877–87.
Gabriele M, Vulto-van Silfhout AT, Germain PL, Vitriolo A, Kumar R, Douglas E, et al. YY1 haploinsufficiency causes an intellectual disability syndrome featuring transcriptional and chromatin dysfunction. Am J Hum Genet. 2017;100:907–25.
Acknowledgements
The authors would like to thank the individuals and their families who participated in this study. We thank Jackie Boyle for her contribution to Family C, Friederike Ruebenstrunk for establishing contact with Family I, and Joop Lavel for technical assistance. The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about. This study was supported by two Dutch NWO VENI grants: OND1312421 to S.G.M.F. and OND1358237 to C.G.P, the European Union grant QLG3-CT-2002-01810 (EuroMRX Consortium), the EU FP7 project GENCODYS, grant number 241995, Australian NHMRC grants 1091593 and 1041920 to J.G., and the European Commission via its Erasmus Joint Doctoral programme 2013-0040 to M.K. C.G. is a grantee of a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Frints, S.G.M., Ozanturk, A., Rodríguez Criado, G. et al. Pathogenic variants in E3 ubiquitin ligase RLIM/RNF12 lead to a syndromic X-linked intellectual disability and behavior disorder. Mol Psychiatry 24, 1748–1768 (2019). https://doi.org/10.1038/s41380-018-0065-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-018-0065-x
This article is cited by
-
Ubiquitin ligases: guardians of mammalian development
Nature Reviews Molecular Cell Biology (2022)
-
Application of exome sequencing for prenatal diagnosis of fetal structural anomalies: clinical experience and lessons learned from a cohort of 1618 fetuses
Genome Medicine (2022)
-
A novel RLIM/RNF12 variant disrupts protein stability and function to cause severe Tonne–Kalscheuer syndrome
Scientific Reports (2021)