Abstract
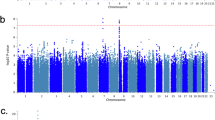
Insomnia is a worldwide problem with substantial deleterious health effects. Twin studies have shown a heritable basis for various sleep-related traits, including insomnia, but robust genetic risk variants have just recently begun to be identified. We conducted genome-wide association studies (GWAS) of soldiers in the Army Study To Assess Risk and Resilience in Servicemembers (STARRS). GWAS were carried out separately for each ancestral group (EUR, AFR, LAT) using logistic regression for each of the STARRS component studies (including 3,237 cases and 14,414 controls), and then meta-analysis was conducted across studies and ancestral groups. Heritability (SNP-based) for lifetime insomnia disorder was significant (h2g = 0.115, p = 1.78 × 10−4 in EUR). A meta-analysis including three ancestral groups and three study cohorts revealed a genome-wide significant locus on Chr 7 (q11.22) (top SNP rs186736700, OR = 0.607, p = 4.88 × 10−9) and a genome-wide significant gene-based association (p = 7.61 × 10−7) in EUR for RFX3 on Chr 9. Polygenic risk for sleeplessness/insomnia severity in UK Biobank was significantly positively associated with likelihood of insomnia disorder in STARRS. Genetic contributions to insomnia disorder in STARRS were significantly positively correlated with major depressive disorder (rg = 0.44, se = 0.22, p = 0.047) and type 2 diabetes (rg = 0.43, se = 0.20, p = 0.037), and negatively with morningness chronotype (rg = −0.34, se = 0.17, p = 0.039) and subjective well being (rg = -0.59, se = 0.23, p = 0.009) in external datasets. Insomnia associated loci may contribute to the genetic risk underlying a range of health conditions including psychiatric disorders and metabolic disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Roth T, Coulouvrat C, Hajak G, Lakoma MD, Sampson NA, Shahly V, et al. Prevalence and perceived health associated with insomnia based on DSM-IV-TR; International Statistical Classification of Diseases and Related Health Problems, Tenth Revision; and Research Diagnostic Criteria/International Classification of Sleep Disorders, Second Edition criteria: results from the America Insomnia Survey. Biol Psychiatry. 2011;69:592–600.
Cunnington D, Junge MF, Fernando AT. Insomnia: prevalence, consequences and effective treatment. Med J Aust. 2013;199:S36–40.
Bramoweth AD, Germain A. Deployment-related insomnia in military personnel and veterans. Curr Psychiatry Rep. 2013;15:401.
Taylor DJ, Pruiksma KE, Hale WJ, Kelly K, Maurer D, Peterson AL, et al. Prevalence, correlates, and predictors of Insomnia in the US Army prior to deployment. Sleep. 2016;39:1795–806.
Krystal JH, Pietrzak RH, Rosenheck RA, Cramer JA, Vessicchio J, Jones KM, et al. Sleep disturbance in chronic military-related PTSD: clinical impact and response to adjunctive risperidone in the Veterans Affairs cooperative study #504. J Clin Psychiatry. 2016;77:483–91.
Morin CM, Drake CL, Harvey AG, Krystal AD, Manber R, Riemann D, et al. Insomnia disorder. Nat Rev Dis Prim. 2015;1:15026.
Jackson CL, Redline S, Emmons KM. Sleep as a potential fundamental contributor to disparities in cardiovascular health. Annu Rev Public Health. 2015;36:417–40.
Anothaisintawee T, Reutrakul S, Van Cauter E, Thakkinstian A. Sleep disturbances compared to traditional risk factors for diabetes development: systematic review and meta-analysis. Sleep Med Rev. 2016;30:11–24.
Fernandez-Mendoza J, Vgontzas AN. Insomnia and its impact on physical and mental health. Curr Psychiatry Rep. 2013;15:418.
Wentworth BA, Stein MB, Redwine LS, Xue Y, Taub PR, Clopton P, et al. Post-traumatic stress disorder: a fast track to premature cardiovascular disease? Cardiol Rev. 2013;21:16–22.
Pollard HB, Shivakumar C, Starr J, Eidelman O, Jacobowitz DM, Dalgard CL, et al. “Soldier’s Heart”: a genetic basis for elevated cardiovascular disease risk associated with post-traumatic stress disorder. Front Mol Neurosci. 2016;9:87.
Wing YK, Zhang J, Lam SP, Li SX, Tang NL, Lai KY, et al. Familial aggregation and heritability of insomnia in a community-based study. Sleep Med. 2012;13:985–90.
Heath AC, Kendler KS, Eaves LJ, Martin NG. Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep. 1990;13:318–35.
Hublin C, Partinen M, Koskenvuo M, Kaprio J. Heritability and mortality risk of insomnia-related symptoms: a genetic epidemiologic study in a population-based twin cohort. Sleep. 2011;34:957–64.
Drake CL, Friedman NP, Wright KP Jr., Roth T. Sleep reactivity and insomnia: genetic and environmental influences. Sleep. 2011;34:1179–88.
Lind MJ, Aggen SH, Kirkpatrick RM, Kendler KS, Amstadter AB. A Longitudinal Twin Study of Insomnia Symptoms in Adults. Sleep. 2015;38:1423–30.
Scheinfeldt LB, Gharani N, Kasper RS, Schmidlen TJ, Gordon ES, Jarvis JP, et al. Using the Coriell personalized medicine collaborative data to conduct a genome-wide association study of sleep duration. Am J Med Genet B Neuropsychiatr Genet. 2015;168:697–705.
Byrne EM, Gehrman PR, Medland SE, Nyholt DR, Heath AC, Madden PA, et al. A genome-wide association study of sleep habits and insomnia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:439–51.
Gottlieb DJ, Hek K, Chen TH, Watson NF, Eiriksdottir G, Byrne EM, et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry. 2015;20:1232–9.
Jones SE, Tyrrell J, Wood AR, Beaumont RN, Ruth KS, Tuke MA, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12:e1006125.
Lane JM, Liang J, Vlasac I, Anderson SG, Bechtold DA, Bowden J, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49:274–81.
Hammerschlag AR, Stringer S, de Leeuw CA, Sniekers S, Taskesen E, Watanabe K, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49:1584–92. [Epub ahead of print]
Van Someren EJ, Cirelli C, Dijk DJ, Van Cauter E, Schwartz S, Chee MW. Disrupted sleep: from molecules to cognition. J Neurosci. 2015;35:13889–95.
Gehrman PR, Keenan BT, Byrne EM, Pack AI. Genetics of sleep disorders. Psychiatr Clin North Am. 2015;38:667–81.
Lind MJ, Gehrman PR Genetic pathways to Insomnia. Brain Sci 2016;6:64; https://doi.org/10.3390/brainsci6040064.
Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–9.
Ursano RJ, Colpe LJ, Heeringa SG, Kessler RC, Schoenbaum M, Stein MB, et al. The Army study to assess risk and resilience in servicemembers (Army STARRS). Psychiatry. 2014;77:107–19.
Kessler RC, Ustun TB. The World Mental Health (WMH) Survey Initiative Version of the World Health Organization (WHO) Composite International Diagnostic Interview (CIDI). Int J Methods Psychiatr Res. 2004;13:93–121.
Kessler RC, Coulouvrat C, Hajak G, Lakoma MD, Roth T, Sampson N, et al. Reliability and validity of the brief insomnia questionnaire in the America insomnia survey. Sleep. 2010;33:1539–49.
Stein MB, Chen CY, Ursano RJ, Cai T, Gelernter J, Heeringa SG, et al. Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of US Army Soldiers. JAMA Psychiatry. 2016;73:695–704.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee SH, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–20.
Chen MH, Pan TL, Li CT, Lin WC, Chen YS, Lee YC, et al. Risk of stroke among patients with post-traumatic stress disorder: nationwide longitudinal study. Br J Psychiatry. 2015;206:302–7.
Major Depressive Disorder Working Group of the Psychiatric GC, Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511.
Psychiatric Genomics Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83.
van den Berg SM, de Moor MH, McGue M, Pettersson E, Terracciano A, Verweij KJ, et al. Harmonization of Neuroticism and Extraversion phenotypes across inventories and cohorts in the Genetics of Personality Consortium: an application of item response theory. Behav Genet. 2014;44:295–313.
Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–33.
Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90.
Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. FUMA: functional mapping and annotation of genetic associations. Nat Commun. 2017;8:1826.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Lind MJ, Hawn SE, Sheerin CM, Aggen SH, Kirkpatrick RM, Kendler KS, et al. An examination of the etiologic overlap between the genetic and environmental influences on insomnia and common psychopathology. Depress Anxiety. 2017;34:453–62.
Byrne EM, Gehrman PR, Trzaskowski M, Tiemeier H, Pack AI. Genetic correlation analysis suggests association between increased self-reported sleep duration in adults and schizophrenia and type 2 diabetes. Sleep. 2016;39:1853–7.
Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108:7119–24.
Chaudhary NS, Kampman KM, Kranzler HR, Grandner MA, Debbarma S, Chakravorty S. Insomnia in alcohol dependent subjects is associated with greater psychosocial problem severity. Addict Behav. 2015;50:165–72.
Benadiba C, Magnani D, Niquille M, Morle L, Valloton D, Nawabi H, et al. The ciliogenic transcription factor RFX3 regulates early midline distribution of guidepost neurons required for corpus callosum development. PLoS Genet. 2012;8:e1002606.
Magnani D, Morle L, Hasenpusch-Theil K, Paschaki M, Jacoby M, Schurmans S, et al. The ciliogenic transcription factor Rfx3 is required for the formation of the thalamocortical tract by regulating the patterning of prethalamus and ventral telencephalon. Hum Mol Genet. 2015;24:2578–93.
Contreras D, Destexhe A, Sejnowski TJ, Steriade M. Control of spatiotemporal coherence of a thalamic oscillation by corticothalamic feedback. Science. 1996;274:771–4.
Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, et al. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12:1373–80.
Wan G, Gomez-Casati ME, Gigliello AR, Liberman MC, Corfas G. Neurotrophin-3 regulates ribbon synapse density in the cochlea and induces synapse regeneration after acoustic trauma. Elife 2014;3, https://doi.org/10.7554/eLife.03564.
Moller-Levet CS, Archer SN, Bucca G, Laing EE, Slak A, Kabiljo R, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110:E1132–1141.
Kato Y, Kawamoto T, Fujimoto K, Noshiro M. DEC1/STRA13/SHARP2 and DEC2/SHARP1 coordinate physiological processes, including circadian rhythms in response to environmental stimuli. Curr Top Dev Biol. 2014;110:339–72.
He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL Jr, et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–70.
Pinggera A, Striessnig J. Cav 1.3 (CACNA1D) L-type Ca2+ channel dysfunction in CNS disorders. J Physiol. 2016;594:5839–49.
Ross J, Gedvilaite E, Badner JA, Erdman C, Baird L, Matsunami N, et al. A rare variant in CACNA1D segregates with 7 bipolar i disorder cases in a large pedigree. Mol Neuropsychiatry. 2016;2:145–50.
Amare AT, Schubert KO, Klingler-Hoffmann M, Cohen-Woods S, Baune BT. The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome wide and candidate gene studies. Transl Psychiatry. 2017;7:e1007.
Ollila HM, Kettunen J, Pietilainen O, Aho V, Silander K, Kronholm E, et al. Genome-wide association study of sleep duration in the Finnish population. J Sleep Res. 2014;23:609–18.
Manoach DS, Pan JQ, Purcell SM, Stickgold R. Reduced sleep spindles in schizophrenia: a treatable endophenotype that links risk genes to impaired cognition? Biol Psychiatry. 2016;80:599–608.
Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with the general population. Am J Epidemiol. 2017;186:1026–34.
Yung G, Lin X. Validity of using ad hoc methods to analyze secondary traits in case-control association studies. Genet Epidemiol. 2016;40:732–43.
Acknowledgements
Funding
Army STARRS was sponsored by the Department of the Army and funded under cooperative agreement number U01MH087981 (2009-2015) with the National Institutes of Health, National Institute of Mental Health (NIH/NIMH). Subsequently, STARRS-LS was sponsored and funded by the Department of Defense (USUHS grant number HU0001-15-2-0004). The contents are solely the responsibility of the authors and do not necessarily represent the views of the Department of Health and Human Services, NIMH, the Department of the Army, or the Department of Defense. Access to Data and Data Analysis: Murray B. Stein MD, MPH had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Drs. Stein, Chen, Jain, McCarthy, and Ripke, as well as Ms. He and Ms. Sun, conducted and are jointly responsible for the data analysis. The Army STARRS Team consists of: Co-Principal Investigators: Robert J. Ursano, MD (Uniformed Services University of the Health Sciences) and Murray B. Stein, MD, MPH (University of California San Diego and VA San Diego Healthcare System). Site Principal Investigators: Steven Heeringa, PhD (University of Michigan), James Wagner, PhD (University of Michigan) and Ronald C. Kessler, PhD (Harvard Medical School). Army liaison/consultant: Kenneth Cox, MD, MPH (USAPHC (Provisional)). Other team members: Pablo A. Aliaga, MS (Uniformed Services University of the Health Sciences); COL David M. Benedek, MD (Uniformed Services University of the Health Sciences); Susan Borja, PhD (NIMH); Tianxi Cai, ScD (Harvard School of Public Health); Laura Campbell-Sills, PhD (University of California San Diego); Carol S. Fullerton, PhD (Uniformed Services University of the Health Sciences); Nancy Gebler, MA (University of Michigan); Robert K. Gifford, PhD (Uniformed Services University of the Health Sciences); Paul E. Hurwitz, MPH (Uniformed Services University of the Health Sciences); Kevin Jensen, PhD (Yale University); Kristen Jepsen, PhD (University of California San Diego); Tzu-Cheg Kao, PhD (Uniformed Services University of the Health Sciences); Lisa Lewandowski-Romps, PhD (University of Michigan); Holly Herberman Mash, PhD (Uniformed Services University of the Health Sciences); James E. McCarroll, PhD, MPH (Uniformed Services University of the Health Sciences); Colter Mitchell, PhD (University of Michigan); James A. Naifeh, PhD (Uniformed Services University of the Health Sciences); Tsz Hin Hinz Ng, MPH (Uniformed Services University of the Health Sciences); Caroline Nievergelt, PhD (University of California San Diego); Nancy A. Sampson, BA (Harvard Medical School); CDR Patcho Santiago, MD, MPH (Uniformed Services University of the Health Sciences); Ronen Segman, MD (Hadassah University Hospital, Israel); Alan M. Zaslavsky, PhD (Harvard Medical School); and Lei Zhang, MD (Uniformed Services University of the Health Sciences).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr. M.B.S. has in the past three years been a consultant for Actelion, Aptinyx, Dart Neuroscience, Healthcare Management Technologies, Janssen, Neurocrine Biosciences, Oxeia Biopharmaceuticals, Pfizer, and Resilience Therapeutics. Dr. M.B.S. owns founders shares and stock options in Resilience Therapeutics and has stock options in Oxeia Biopharmaceticals. Dr. J.W.S. is an unpaid member of the Scientific Advisory Board of PsyBrain, Inc. In the past 3 years, Dr. R.C.K. has been a consultant for Hoffman-La Roche, Inc., Johnson & Johnson Wellness and Prevention, and Sanofi-Aventis Groupe. Dr. R.C.K. has served on advisory boards for Mensante Corporation, Plus One Health Management, Lake Nona Institute, and US Preventive Medicine. Dr. R.C.K. owns 25% share in DataStat, Inc. The remaining authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Stein, M.B., McCarthy, M.J., Chen, CY. et al. Genome-wide analysis of insomnia disorder. Mol Psychiatry 23, 2238–2250 (2018). https://doi.org/10.1038/s41380-018-0033-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-018-0033-5
This article is cited by
-
Nilpotent Singularities and Periodic Perturbation of a \(GI\beta \) Model: A Pathway to Glucose Disorder
Journal of Nonlinear Science (2023)
-
Identification of six genes associated with COVID-19-related circadian rhythm dysfunction by integrated bioinformatic analysis
Functional & Integrative Genomics (2023)
-
Towards organism-level systems biology by next-generation genetics and whole-organ cell profiling
Biophysical Reviews (2021)
-
Identifying novel genetic variants for brain amyloid deposition: a genome-wide association study in the Korean population
Alzheimer's Research & Therapy (2021)
-
Genome-wide association analysis of insomnia using data from Partners Biobank
Scientific Reports (2020)