Abstract
SMARCA4-deficient undifferentiated malignant neoplasms (SD-UMN) comprise a group of aggressive tumors with epithelioid morphology that are characterized by loss of function of SMARCA4, a component of the SWI/SNF chromatin remodeling complex. SD-UMN was first recognized in the thoracic cavity but is now appreciated to occur at multiple anatomic sites. A notable exception has been skin. Here we report the first two cases of primary cutaneous SD-UMN and compare their features to a cohort of eight visceral cases arising in lung, gastrointestinal tract, and gallbladder. Evidence for a bona fide cutaneous origin included extensive clinical, radiologic, and serologic analyses that failed to identify a metastatic source as well as the molecular identification of a UV-associated mutational pattern. The cutaneous cases showed strikingly similar morphologic, immunohistochemical, and molecular features to the visceral cases, strongly suggesting that they belong to this family of tumors. In addition to biallelic inactivation of SMARCA4, both cutaneous tumors also showed biallelic inactivation of TP53 and CDKN2A, findings which also appear common in visceral cases. One patient died of disease at 18 months after diagnosis, consistent with the aggressive nature of this tumor. Our results expand the anatomic spectrum of SD-UMN, adding this entity to an already challenging differential diagnosis that includes melanoma, squamous cell carcinoma, Merkel cell carcinoma, epithelioid sarcoma, and others. Given the potentially aggressive nature of SD-UMN, the timely and accurate diagnosis of this entity may have implications for prognosis and therapy.
Similar content being viewed by others
Introduction
SMARCA4-deficient undifferentiated malignant neoplasms (SD-UMN) encompass a family of malignant epithelioid tumors often associated with poor clinical outcomes. The defining feature is the presence of inactivating SMARCA4 mutations, with the resultant loss of protein expression being detectable by immunohistochemistry1,2. SMARCA4 (also known as BRG1) is an ATPase which comprises one of the two catalytic subunits of the SWItch/Sucrose Nonfermentable (SWI/SNF) chromatin remodeling complex (the other being SMARCA2; reviewed in Mittal et al.3). The SWI/SNF complex is involved in transcriptional regulation and cellular proliferation, and loss of function of SMARCA4 is a well-established oncogenic driver both in experimental models and in various cancer types4,5.
Malignancies characterized by SMARCA4 deficiency have a wide anatomic distribution and include tumors of the lung, gastrointestinal tract, sinonasal tract, and gynecologic tract6,7,8,9,10,11,12. Regardless of the site of origin, SD-UMN are characterized by common histopathologic characteristics, including an undifferentiated epithelioid large cell or rhabdoid cytomorphology. Atypical mitoses, apoptotic debris, and tumor necrosis are also common findings. Their relatively undifferentiated appearance can make the diagnosis challenging, and raises consideration of high-grade epithelial, mesenchymal, endothelial, and melanocytic neoplasms in the differential diagnosis.
One notable exception within the recognized anatomic spectrum of SD-UMN is skin. Here we describe the first cases of primary cutaneous SD-UMN and report their genomic characterization by next generation DNA sequencing. We compare the clinicopathologic, immunohistochemical, and molecular features of these primary cutaneous tumors to SD-UMN arising at visceral sites. Our findings have important implications for the diagnosis of undifferentiated epithelioid malignancies in the skin.
Materials and methods
Case selection
Approval was obtained for this study from our Institutional Review Board. Two cases of primary cutaneous SD-UMN were identified at our institution between 2019 and 2021. In an effort to identify more cases, our institutional files were searched for cutaneous cases with terms including “epithelioid”, “rhabdoid”, “poorly differentiated carcinoma/neoplasm”, and “undifferentiated carcinoma/neoplasm”; however, no additional cutaneous cases have been identified to date. Additional representative cases of SD-UMN at visceral sites were retrospectively selected from our institutional files. These eight visceral cases were selected based upon the availability of slides for review and the completion of relevant molecular studies. No cases were subsequently excluded after initial selection.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on 4 µm-thick formalin fixed, paraffin-embedded tissue sections. SMARCA4 IHC was performed following citrate buffer pressure cooker epitope retrieval (Target Retrieval Solution, pH 6.1; Dako, Carpinteria, CA) using a rabbit anti-SMARCA4 (BRG1) monoclonal antibody (1:50 dilution, 40 min incubation, clone EPR3912; Abcam, Cambridge, MA). Sox-2 IHC was performed on 4 µm-thick formalin-fixed paraffin-embedded tissue sections following citrate buffer pressure cooker epitope retrieval (Target Retrieval Solution, pH 6.1; Dako) using a rabbit anti-Sox-2 monoclonal antibody (1:75 dilution, 40 min incubation; CLONE D6D9; Cell Signaling Techonology, Danvers, MA). Pan-keratin IHC was performed on 4 µm-thick formalin-fixed paraffin-embedded tissue sections following protease enzyme digestion epitope retrieval using a mouse anti-pan-keratin monoclonal antibody (clone MNF116, 1:300 dilution, 40 min incubation; Agilent/Dako, Santa Clara, CA). EnVision plus detection system (Dako Link 48) was used for all antibodies.
Molecular profiling
Two cases of primary cutaneous SD-UMN were analyzed using a next-generation DNA sequencing platform (Oncopanel), as previously described13,14. Cases met adequacy requirements of at least 20% tumor cells in specimens measuring at least 3 mm in greatest linear dimension. Eight additional cases of primary visceral SD-UMN were analyzed in parallel using this method. The Oncopanel assay surveys exonic DNA sequences of 447 cancer genes and can identify alterations including insertions, deletions and substitutions. In addition, 191 regions across 60 genes are interrogated for the detection of chromosomal rearrangements. A complete list of genes included can be found in supplemental table S1. Regions harboring single nucleotide polymorphisms (SNPs), spaced approximately every 4 MB across the genome are also captured and sequenced to facilitate assessment of copy number variations. Notably the Oncopanel assay includes SMARCA4 (BRG1), the related gene SMARCB1 (INI1), and diverse driver genes involved in the pathogenesis of entities in the histopathologic differential diagnosis such as melanoma, squamous cell carcinoma, and cutaneous metastases (such as from lung and gastrointestinal primaries). Additionally, for tumors with 16 or more mutations, mutational signature analysis is performed based upon the pattern of nucleotide substitutions, allowing recognition of signatures associated with DNA damage due to ultraviolet light (UVA) exposure, tobacco smoke exposure, prior treatment with alkylating agents (including temozolomide), impaired POLE DNA polymerase function, and APOBEC (Apolipoprotein B mRNA Editing Catalytic Polypeptide-like) enzyme dysregulation. The Oncopanel mutational signature detection tool is based upon previously published signatures derived from whole exome sequencing data15 and was subsequently refined by training on targeted exome sequencing data16. The reported mutational patterns reflect those observed in vitro following exposure to relevant mutagens. The presence of these signatures was further validated against the clinicopathologic features in 740 Oncopanel samples including origin at a sun-exposed site, smoking history, prior treatment with temozolomide, concurrent POLE hotspot mutation, or MMR deficiency, as detected by Oncopanel.
Results
Clinical features of cutaneous and visceral SMARCA4-deficient undifferentiated malignant neoplasms
The clinical features of all ten cases are summarized in Table 1. Both cases of primary cutaneous SD-UMN were received in consultation at our institution with a referring diagnosis of Merkel cell carcinoma. Both patients were men (84 and 70 years old). Patient 1 presented with a single papule on the left neck, clinically concerning for a basal cell carcinoma. Clinical and radiologic examination, including a full body PET scan, did not reveal evidence of visceral or other metastatic disease. Serologic testing for Merkel cell carcinoma-associated antibodies was negative. A wide excision was performed but tumor recurred locally within one year. The patient underwent radiation therapy, but within 4 months new lesions arose in the radiation field. Despite yet another excision, the disease progressed with continued local recurrences and the patient was referred to hospice care where he died approximately 18 months following initial diagnosis. Patient 2 developed a rapidly growing keratotic papule on the right cheek. The patient had a history of melanoma, also on the right cheek, 10 years previously and for which he underwent wide local excision and negative sentinel lymph node biopsy. The excision of his SD-UMN did not show a scar, confirming that the two tumors were at distinct sites on the cheek. Extensive clinical and radiologic evaluation, including computed tomography (CT) of the thorax, abdomen, and pelvis, as well as full body PET scan, failed to reveal a metastatic source or other foci of disease. Serologic testing for Merkel cell carcinoma-associated antibodies was again negative. The patient underwent complete excision of the biopsy site with minimal residual tumor. One year following diagnosis the patient remained recurrence-free. Repeat CT scans of the head, neck, and have been negative to date, and the patient remains under close surveillance with repeat imaging every six months.
Patients with primary visceral SMARCA4-deficient undifferentiated malignant neoplasms (n = 8: female = 4, male = 4) ranged in age from 42–71 with a median age of 68. Primary tumor sites included lung (n = 4), esophagus (n = 2), rectum (n = 1) and gallbladder (n = 1). Five patients had progressive disease despite treatment and died between 2–19 months post-diagnosis. Two patients remain disease-free at 15 months and 5 years post-diagnosis, respectively. Follow-up data was not available for the remaining case. Further details including treatment and follow-up interval information are summarized in Table 1.
Morphologic and immunohistochemical features of cutaneous and visceral SMARCA4-deficient undifferentiated malignant neoplasms
The morphologic features of the primary cutaneous SD-UMN were highly similar to those of visceral origin. All of the tumors were composed of medium to large epithelioid cells with prominent nucleoli (sometimes multiple), a fine chromatin pattern, and thickened and irregular nuclear membranes (Fig. 1). The cells contained moderate amounts of amphophilic cytoplasm and some displayed areas of rhabdoid cytomorphology. Mitotic activity and apoptotic debris were frequent findings. For the cutaneous tumors, there was no intraepidermal component. Rather the tumors were primarily located in the dermis (Fig. 1) and showed sheet-like growth with some areas showing a more nested pattern. The tumor border was relatively well-circumscribed for case 1; this feature was difficult to assess in case 2 due to the small size of the sample. Subsequent wide excision of the tumors revealed focal extension of tumor into the subcutis in case 1 and only focal residual dermal involvement in case 2. Perineural invasion was identified in case 1.
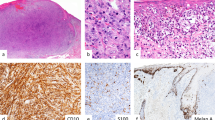
Case 1 A, B shows a poorly differentiated malignant neoplasm with sheet-like growth occupying the dermis after component:...without an in-situ component (A). Higher magnification shows that the tumor is composed of pleomorphic epithelioid cells with amphophilic cytoplasm and variably prominent nucleoli. Mitoses are easily identified, and apoptotic cells are present (B). Case 2 C, D shows a remarkably similar morphology. A low power image shows a dermally-based tumor lacking an in-situ component with a more nested architecture (C). Higher magnification shows that the tumor is composed of pleomorphic epithelioid cells with variably prominent nucleoli and focal rhabdoid morphology (arrow, D). Mitoses are frequent. (All images show H&E-stained slides. Panels A and C, 100 x magnification. Panels B and D, 400 x magnification).
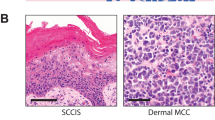
Complete loss of SMARCA4 immunohistochemical expression was demonstrated in all cutaneous and visceral cases (Figs. 2 and 3). Nuclear expression was retained in neighboring non-neoplastic cells. The primary cutaneous tumors additionally expressed pan-keratin and Sox2 (Fig. 2). Given the broad differential diagnosis that included poorly differentiated squamous cell carcinoma, melanoma, Merkel cell carcinoma, NUT midline carcinoma, and other epithelioid malignancies, a wide array of additional immunohistochemical studies were performed but all were negative including p63, INSM1, cytokeratin 7 (CK7), CK20, ERG, CD34, MCPyV (Merkel cell polyoma virus large T antigen), NUT (nuclear protein in testis), Sox-10, S100, Melan-A, and HMB45. INI1 expression was retained, arguing against an epithelioid sarcoma.
Case 1 is depicted in the top row A–C, and case 2 is depicted in the bottom row D–F. Both cases showed complete loss of staining of SMARCA4 (A, D; note retained nuclear expression in non-neoplastic cells). Primary cutaneous SMARCA4-deficient malignant neoplasms also express pan-keratin (B, E) and Sox-2 (C, F). E shows small volume of tumor due to loss of tissue on deeper levels. All images depict 200x magnification.
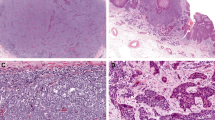
A shows a primary lung SMARCA4-deficient undifferentiated malignant neoplasm composed of pleomorphic epithelioid cells, frequent mitoses, and apoptotic debris. This tumor shows complete loss of SMARCA4 expression (with retention of expression in non-noeplastic cells) (B). A primary rectal SMARCA4-deficient undifferentiated malignant neoplasm shows classic epithelioid morphology with variably prominent nucleoli (C). This tumor shows complete loss of SMARCA4 expression (with retention of expression in non-neoplastic cells). Interestingly, the rectal epithelium also shows loss of SMARCA4 expression, which is of uncertain significance but suggests that it may represent neoplastic or pre-neoplastic epithelium (D). All images depict 200x magnification.
Tumors arising in the lung showed variable keratin and Sox2 expression and were negative for TTF-1, napsin-A, p40, INSM1, and NUT. Tumors arising in the esophagus showed variable keratin expression and retention of INI1; they were negative for CDX-2, p63, INSM1, and S100 expression. The rectal primary showed keratin expression, focal SATB2 expression and was negative for CDX-2. The gallbladder primary was negative for Sox10 and INSM1 expression.
Molecular characteristics of cutaneous and visceral SMARCA4-deficient undifferentiated malignant neoplasms
Next-generation DNA sequencing was performed on all ten cases, with results summarized in Table 2. Cutaneous SD-UMN case 1 harbored two nonsense variants in SMARCA4, c.535 C > T and c.4531 A > T, which result in premature termination at Q179 and K1511, respectively. Neither mutation has been previously reported in COSMIC (Catalogue of Somatic Mutations in Cancer)17. While the sequencing data cannot distinguish between the two mutations occurring in the same allele or in different alleles, the complete loss of SMARCA4 expression (Fig. 2) supports biallelic inactivation in this patient. The second cutaneous SD-UMN patient also showed two SMARCA4 mutations, again consistent with biallelic inactivation. The first was a known pathogenic variant (c.1492 C > T) which results in premature termination at Q498 while the second was a splice site variant predicted to lead to a frameshift alteration (c.1943_1943 + 1delinsAA). This patient additionally showed single copy deletion of the related gene INI-1 (also known as SMARCB1). This is interesting because INI-1 is also a member of the SWI/SNF complex. However, INI-1 expression was retained in this patient’s tumor, and so the pathologic significance of this finding remains uncertain. Importantly, both cutaneous tumors harbored an ultraviolet radiation (UV)-associated mutational signature, which supports their cutaneous origin and which was absent in all visceral cases.
Two other mutational events were shared between the two skin tumors. The first was apparent biallelic inactivation of CDKN2A (p16). In case 1, this occurred through two-copy deletion of the locus (Fig. 4), while in case 2 this occurred through loss of one allele combined with the acquisition of a known pathogenic mutation in the other allele (c.341_342delinsTT; p.P114L). The second shared mutational event was apparent biallelic loss of TP53. Case 1 showed two known pathogenic mutations (c.772 G > A; p.E258K and c.833 C > T; p.P278L) while case 2 showed loss of one allele at chromosome 17p13.1 coupled with three known pathogenic mutations in the remaining allele (c.783-1 G > A, c.796_797delinsAA;p.G266K, and c.836_837delinsAA; p.G279E).
Magenta and gold dots indicate the log2 ratio of sample copy number relative to a pooled normal at the level of individual exons and selected introns for each of the targeted genes. Green dots indicate a copy number other than neutral as called by automated algorithms. Pale blue tracing shows the percent guanine and cytosine (GC) in the targeted region. The green line shows the median log2 ratio for all samples on a plate; the black line shows the ratio for the specific case.
All 8 visceral SD-UMN cases also showed loss of function of SMARCA4 aberrations. Interestingly, this occurred through a variety of mechanisms including mutation, two copy loss of the locus, and mutation combined with single copy loss of the locus (Table 2). Nearly all cases from visceral sites harbored CDKN2A loss-of-function aberrations (7/8 cases, 88%), suggesting that this may be a characteristic feature of this tumor family. TP53 loss-of-function aberrations were also prominent (4/8 cases, 50%), although less uniformly so than CDKN2A.
There were some additional findings of interest. One of the lung tumors (case 2) showed known pathogenic mutations in KRAS, STK11, and KEAP1, which are recognized mutations in lung carcinomas. A different lung tumor (case 3) showed single copy loss of INI-1 (SMARCB1), similar to one of the cutaneous cases. Interestingly, none of the visceral cases, including the four lung cases, showed a tobacco-associated mutational profile.
A variety of other mutations and copy number changes were present within all 10 cases, but we did not identify other clear patterns of shared or recurrent aberrations.
Discussion
Here we report the first cases of primary cutaneous SD-UMN. Our assignment of these cases as bona fide primary tumors is based on extensive clinical, radiologic, and serologic analyses which failed to reveal any metastatic source, coupled with the molecular identification of a UV-associated mutational profile. This latter finding strongly supports a cutaneous origin and was not seen in any of the visceral tumors. We considered the possibility of metastasis from a regressed visceral primary. This sort of spontaneous regression is not well described for visceral SD-UMN and, in any case, the presence of a UV signature in these tumors argues against this model. The morphologic, immunohistochemical, and molecular features of cutaneous SD-UMN are strikingly similar to its visceral counterparts, arguing that they belong in the overall family of SD-UMN. Thus, our work expands the anatomic spectrum of this potentially aggressive tumor with important implications for diagnosis, prognosis, and therapy.
The differential diagnosis for poorly differentiated epithelioid malignancies in the skin is broad and challenging. The main considerations included poorly differentiated squamous cell carcinoma, melanoma, Merkel cell carcinoma, NUT midline carcinoma, epithelioid sarcoma, and others18,19. Our work highlights the importance of a broad immunohistochemical work-up which, in principle, should be able to distinguish between these entities through the identification of SMARCA4 loss coupled with exclusion of the other entities (e.g. lack of melanocytic or squamous markers, lack of CK20 and MCPyV, retention of INI-1, absence of NUT, and so on). Accordingly, when dealing with poorly differentiated epithelioid or rhabdoid dermal neoplasms which lack lineage-specific markers, evaluation of SMARCA4 expression may be helpful before rendering a diagnosis of poorly differentiated carcinoma/malignant neoplasm. Molecular analysis may be useful in further confirming the diagnosis and in excluding the known driver mutations of other entities, although most diagnoses should be possible by morphology and immunohistochemistry, coupled with strong clinico-pathologic correlation. Care should also be taken, primarily through clinical correlation, to exclude metastatic SD-UMN to the skin from an underlying visceral source, as has been recently documented19. Even within the differential diagnosis discussed above, SD-UMN stands out as being particularly aggressive. As more primary cutaneous SD-UMN cases are identified, it will be important to determine whether they show comparable rates of aggressiveness to their visceral counterparts6,9,20. Some of the entities in the differential diagnosis also have individualized therapeutic approaches, including sentinel node sampling, immunotherapy, and others21,22. Thus, the timely and accurate recognition of SD-UMN will be important to ensure selection of the proper therapeutic approach.
Our data also provide detailed molecular analysis of a relatively large series of SD-UMN from diverse anatomic sites, with a number of interesting findings. First, while loss of SMARCA4 expression is a hallmark of this entity (Figs. 2 and 3), the data reveal multiple molecular pathways to this end, including mutation, copy number loss, and a combination of the two. Indeed, this emphasizes the utility of additionally measuring copy number changes, and not just mutations, in understanding the full genomic profile. There may be other pathways to SMARCA4 inactivation that remain to be discovered. Targeted protein degradation and epigenetic regulation are interesting candidates. A second interesting feature was the presence of recurrent loss-of-function alterations in CDKN2A (9/10 cases, illustrated in Fig. 4) and TP53 (6/10 cases). While these two genes are among the most commonly mutated in cancer, their prominence in this series of SD-UMN suggests that they, along with SMARCA4, may represent a core mutational module for this entity. Indeed, prior studies have found TP53 (5/5 cases) and CDKN2A (2/5 cases) alterations in SMARCA4-deficient thoracic sarcoma23. Interestingly, in contrast, genomic profiling of SMARCA4-deficient uterine sarcoma and small cell carcinoma of the of the ovary, hypercalcemic type (which is defined by biallelic loss of function of SMARCA4), failed to reveal recurrent TP53 or CDKN2A alterations, suggesting that TP53 and CDKN2A alterations may play a more important role in tumors arising outside of the gynecologic tract12,24,25.
As for nomenclature, there has been much discussion about whether this entity represents a carcinoma or a sarcoma6,7. Our understanding of SD-UMN is evolving, as highlighted by the improved characterization of the SMARCA4-deficient group of thoracic neoplasms. Initially described as SMARCA4-deficient thoracic sarcomas6,26,27, recent work now proposes that these thoracic tumors largely represent smoking-related undifferentiated carcinomas rather than sarcomas7. Conversely, the lack of claudin-4 expression and the presence of Sox2 expression in some SMARCA4-deficient neoplasms with epithelioid morphology has been proposed by some authors to support a mesenchymal origin for these tumors28. We do not know the cell type of origin for the cutaneous SD-UMN and have therefore used the neutral term “malignant neoplasm” for now.
In summary, we report here the first two cases of primary cutaneous SD-UMN and show that their morphologic, immunohistochemical, and molecular features are highly similar to their visceral counterparts. Increased awareness of this entity should improve both diagnosis and clinical care. We expect that the identification of additional cases will clarify important aspects of this tumor including its true incidence, natural history, and pathophysiologic mechanisms.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ramos, P, Karnezis, AN, Craig, DW, Sekulic, A, Russell, ML, Hendricks, WPD, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 46:427–429 (2014).
Karanian-Philippe, M, Velasco, V, Longy, M, Floquet, A, Arnould, L, Coindre, J-M, et al. SMARCA4 (BRG1) loss of expression is a useful marker for the diagnosis of ovarian small cell carcinoma of the hypercalcemic type (ovarian rhabdoid tumor): a comprehensive analysis of 116 rare gynecologic tumors, 9 soft tissue tumors, and 9 melanomas. Am J Surg Pathol. 39:1197–1205 (2015).
Mittal, P, Roberts, CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol. 17:435–448 (2020).
Wong, AK, Shanahan, F, Chen, Y, Lian, L, Ha, P, Hendricks, K, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177 (2000).
Kadoch, C, Hargreaves, DC, Hodges, C, Elias, L, Ho, L, Ranish, J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 45:592-601 (2013).
Le Loarer, F, Watson, S, Pierron, G, de Montpreville, VT, Ballet, S, Firmin, N, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 47:1200-1205 (2015).
Rekhtman, N, Montecalvo, J, Chang, JC, Alex, D, Ptashkin, RN, Ai, N, et al. SMARCA4-deficient thoracic sarcomatoid tumors represent primarily smoking-related undifferentiated carcinomas rather than primary thoracic sarcomas. J Thorac Oncol. 15:231-247 (2020).
Agaimy, A, Daum, O, Märkl, B, Lichtmannegger, I, Michal, M, Hartmann, A. SWI/SNF complex-deficient undifferentiated/rhabdoid carcinomas of the gastrointestinal tract: A series of 13 cases highlighting mutually exclusive loss of SMARCA4 and SMARCA2 and frequent co-inactivation of SMARCB1 and SMARCA2. Am J Surg Pathol. 40:544-553 (2016).
Agaimy, A, Jain, D, Uddin, N, Rooper, LM, Bishop, JA. SMARCA4-deficient sinonasal carcinoma: A series of 10 cases expanding the genetic spectrum of SWI/SNF-driven sinonasal malignancies. Am J Surg Pathol. 44:703-710 (2020).
Jelinic, P, Mueller, JJ, Olvera, N, Dao, F, Scott, SN, Shah, R, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 46:424-426 (2014).
Lin, DI, Allen, JM, Hecht, JL, Killian, JK, Ngo, NT, Edgerly, C, et al. SMARCA4 inactivation defines a subset of undifferentiated uterine sarcomas with rhabdoid and small cell features and germline mutation association. Mod Pathol. 32:1675-1687 (2019).
Kolin, DL, Quick, CM, Dong, F, Fletcher, CDM, Stewart, CJR, Soma, A, et al. SMARCA4-deficient uterine sarcoma and undifferentiated endometrial carcinoma are distinct clinicopathologic entities. Am J Surg Pathol. 44:263-270 (2020).
Russell-Goldman, E, MacConaill, L, Hanna, J. Hedgehog pathway alterations downstream of Patched-1 are common in infundibulocystic basal cell carcinoma. Am J Dermatopathol. 43:266-272 (2021).
Russell-Goldman, E, Lindeman, NI, Laga, AC, Hanna, J. Morphologic, immunohistochemical, and molecular distinction between fibroepithelioma of pinkus and “fenestrated” basal cell carcinoma. Am J Dermatopathol. 42:513-520 (2020).
Alexandrov, L, Nik-Zainal, S, Wedge, D, Aparicio, SAJR, Behjati, S, Biankin, AV, et al. Signatures of mutational processes in human cancer. Nature 500:415–421 (2013).
Zehir, A, Benayed, R, Shah, RH, Syed, A, Middha, S, Kim, HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 23:703–713 (2017).
Tate, JG, Bamford, S, Jubb, HC, Sondka, Z, Beare, DM, Bindal, N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47:D941-D947 (2019).
Carter, CS, Patel, RM. Cutaneous soft tissue tumors: diagnostically disorienting epithelioid tumors that are not epithelial, and other perplexing mesenchymal lesions. Mod Pathol. 33(Suppl 1):66-82 (2020).
Leckey, BD Jr, Selim, MA, Al-Rohil, RN. Cutaneous metastasis of SMARCA4-deficient thoracic sarcoma: A diagnostic dilemma with therapeutic implications. J Cutan Pathol. 47:561-565 (2020).
Chang B, Sheng W, Wang L, Zhu, X, Tan, C, Ni, S, et al. SWI/SNF Complex-deficient undifferentiated carcinoma of the gastrointestinal tract: Clinicopathologic study of 30 cases with an emphasis on variable morphology, immune features, and the prognostic significance of different SMARCA4 and SMARCA2 subunit deficiencies [published online ahead of print, 2021 Nov 23]. Am J Surg Pathol. https://doi.org/10.1097/PAS.0000000000001836 (2021).
Cassler, NM, Merrill, D, Bichakjian, CK, Brownell, I. Merkel cell carcinoma therapeutic update. Curr Treat Options Oncol. 17:36 (2016).
Curti, BD, Faries, MB. Recent advances in the treatment of melanoma. N. Engl J Med. 384:2229-2240 (2021).
Yoshida, A, Kobayashi, E, Kubo, T, Kodaira, M, Motoi, T, Motoi, N, et al. Clinicopathological and molecular characterization of SMARCA4-deficient thoracic sarcomas with comparison to potentially related entities. Mod Pathol. 30:797-809 (2017).
Lin, DI, Chudnovsky, Y, Duggan, B, Zajchowski, D, Greenbowe, J, Ross, JS, et al. Comprehensive genomic profiling reveals inactivating SMARCA4 mutations and low tumor mutational burden in small cell carcinoma of the ovary, hypercalcemic-type. Gynecol Oncol. 147:626-633 (2017).
Auguste, A, Blanc-Durand, F, Deloger, M, Le Formal, A, Bareja, R, Wilkes, DC, et al. Small Cell Carcinoma of the Ovary, Hypercalcemic Type (SCCOHT) beyond SMARCA4 mutations: A comprehensive genomic analysis. Cells 9:1496 (2020).
Sauter, JL, Graham, RP, Larsen, BT, Jenkins, SM, Roden, AC, Boland, JM. SMARCA4-deficient thoracic sarcoma: a distinctive clinicopathological entity with undifferentiated rhabdoid morphology and aggressive behavior. Mod Pathol. 30:1422-1432 (2017).
Perret, R, Chalabreysse, L, Watson, S, Serre, I, Garcia, S, Forest, F, et al. SMARCA4-deficient Thoracic Sarcomas: Clinicopathologic study of 30 cases with an emphasis on their nosology and differential diagnoses. Am J Surg Pathol. 43:455-465 (2019).
Schaefer, IM, Agaimy, A, Fletcher, CD, Hornick, JL. Claudin-4 expression distinguishes SWI/SNF complex-deficient undifferentiated carcinomas from sarcomas. Mod Pathol. 30:539-548 (2017).
Acknowledgements
The authors wish to thank Mei Zheng for assistance with immunohistochemistry, and the staff at the Center for Advanced Molecular Diagnostics (CAMD), Brigham and Women’s Hospital.
Funding
This work was supported by a grant from the Bertarelli Rare Cancers Fund (to J.H.), and by the Department Commitment Fund from the Department of Pathology at Brigham and Women’s Hospital (to E.R.G).
Author information
Authors and Affiliations
Contributions
Concept and design: E.R.G. and J.H. Case contribution: E.R.G. and J.H. Pathology review: E.R.G. and J.H. Molecular analysis and interpretation: L.M., E.R.G. and J.H. Original manuscript draft preparation: E.R.G. and J.H. Review and editing of manuscript: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Cases were included with approval from our Institutional Review Board.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Russell-Goldman, E., MacConaill, L. & Hanna, J. Primary cutaneous SMARCA4-deficient undifferentiated malignant neoplasm: first two cases with clinicopathologic and molecular comparison to eight visceral counterparts. Mod Pathol 35, 1821–1828 (2022). https://doi.org/10.1038/s41379-022-01152-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-022-01152-1