Abstract
Adult-type diffuse gliomas and meningiomas are the most common primary intracranial tumors of the central nervous system. DNA methylation profiling is a novel diagnostic technique increasingly used also in the clinic. Although molecular heterogeneity is well described in these tumors, DNA methylation heterogeneity is less studied. We therefore investigated the intratumor genetic and epigenetic heterogeneity in diffuse gliomas and meningiomas, with focus on potential clinical implications. We further investigated tumor purity as a source for heterogeneity in the tumors. We analyzed genome-wide DNA methylation profiles generated from 126 spatially separated tumor biopsies from 39 diffuse gliomas and meningiomas. Moreover, we evaluated five methods for measurement of tumor purity and investigated intratumor heterogeneity by assessing DNA methylation-based classification, chromosomal copy number alterations and molecular markers. Our results demonstrated homogeneous methylation-based classification of IDH-mutant gliomas and further corroborates subtype heterogeneity in glioblastoma IDH-wildtype and high-grade meningioma patients after excluding samples with low tumor purity. We detected a large number of differentially methylated CpG sites within diffuse gliomas and meningiomas, particularly in tumors of higher grades. The presence of CDKN2A/B homozygous deletion differed in one out of two patients with IDH-mutant astrocytomas, CNS WHO grade 4. We conclude that diffuse gliomas and high-grade meningiomas are characterized by intratumor heterogeneity, which should be considered in clinical diagnostics and in the assessment of methylation-based and molecular markers.
Similar content being viewed by others
Introduction
Diffuse gliomas and meningiomas are the most prevalent intracranial tumors of the central nervous system (CNS) in adults1. Meningiomas are typically benign tumors and can potentially be cured by surgical resection, whereas high-grade meningiomas carry an increased risk of recurrence and need of adjuvant treatment2. Diffuse gliomas, including IDH-mutant glioma (CNS WHO grade 2–4) and glioblastoma IDH-wildtype (CNS WHO grade 4), are infiltrating diseases that are currently incurable3. Tumor heterogeneity is considered one of the contributors to tumor recurrence and treatment failure4. While multiplatform high-throughput analyzes have revealed the cellular and molecular heterogeneity between and within diffuse gliomas and meningiomas5,6,7,8,9,10,11,12,13, the intratumor epigenetic heterogeneity is less investigated.
DNA methylation is a key epigenetic modification controlling several genomic functions, and methylation aberrations have been identified to play an important role in the development of human cancers14,15. Characterization of DNA methylation patterns has led to the identification of biologically and clinically relevant molecular subgroups of CNS tumors5,16,17,18,19 with diagnostic and prognostic potential. On this basis, the current 2021 WHO classification highlights the relevance of employing methylation profiling as a tool for reliable identification of tumor types and subtypes in the routine clinical practice alongside with standard methods20. With presumed increase in clinical use, it is necessary to establish aspects of diagnostic reproducibility, such as robustness of diagnosis throughout the tumor volume. Low level of heterogeneity would imply less risk of surgical sampling bias, a common problem in establishing WHO grade from small tissue specimens21 (e.g., biopsies only). In glioblastoma, WHO (2016) grade IV, we previously demonstrated that intratumor DNA methylation heterogeneity exists, with potential implications for methylation-based classification22.
One factor that may contribute to heterogeneity in DNA methylation profiles is cancer cells and non-cancer cell types intermingled in bulk tumor samples. The presence of normal cells within the tumors could potentially mask methylation signals from the tumor and consequently mislead biological interpretation of the methylation data23,24. Recently, several methods have been developed to estimate the proportion of neoplastic cells in the tumor sample, here denoted as tumor purity, based on DNA methylation array data25,26,27,28,29, but a comprehensive evaluation of these methods in different brain tumor types is limited in the literature.
In this study, we used DNA methylation data and chromosomal copy number data inferred from genome-wide DNA methylation arrays, to investigate heterogeneity across 39 diffuse gliomas and meningiomas. We further compared the performance of different methods for estimation of tumor purity to address tumor purity as a confounding factor in heterogeneity analyses. After excluding samples with low tumor purity, we found homogeneous methylation class families in IDH-mutant gliomas CNS WHO grade 2–4, glioblastomas IDH-wildtype, CNS WHO grade 4 and meningiomas, CNS WHO grade 1–2 but distinct molecular subgroups of glioblastoma IDH-wildtype, CNS WHO grade 4 and high-grade meningioma within some of the tumors. Differences in tumor purity could not explain the observed subtype/subclass heterogeneity in most cases, but could not be ruled out as a factor in a few. We also encountered high number of differentially methylated CpG sites within the tumors, which was associated with CNS WHO grade. Our study demonstrates intratumor heterogeneity in diffuse gliomas and meningiomas, which should be accounted for in clinical diagnostics.
Materials and methods
Patients and sample collection
The patients (≥18 years old) underwent surgical tumor resection of primary or recurrent diffuse lower-grade glioma (as defined by TCGA10) IDH-wildtype/IDH-mutant, WHO (2016) grade II–III, glioblastoma IDH-wildtype/IDH-mutant, WHO (2016) grade IV or high-grade meningioma, WHO (2016) grade II, during 2018–2020 at the neurosurgical department at the Sahlgrenska University hospital (Gothenburg, Sweden). The diagnosis of the patients followed the WHO classification criteria valid at the time of surgery (2016 WHO) and the samples were labeled according to the type of tumor; lower-grade glioma (GU-LGG-X), high-grade glioma (GU-HGG-X) and high-grade meningioma (GU-hgMNG-X), where X represents a serial ID number. Signed informed consent was obtained from all patients in this study. For each patient, three to five tumor tissue biopsies were sourced from different regions of the tumor with a spatial distance of at least 1 cm between the biopsies. Each biopsy was divided into small pieces and cryopreserved for molecular or histological assessments.
DNA isolation and methylation array profiling
DNA extraction from fresh-frozen tumor biopsies, generation and processing of methylation data inferred from the Infinium MethylationEPIC BeadChip array (Illumina, Inc., San Diego, CA, US) as well as the analysis of differentially methylated positions (DMPs) between intratumor biopsy pairs were carried out as previously described22. We used the statistical software R with Rstudio (version 4.0.2) for data analysis. Prediction of the O6-methylguanine-DNA methyltransferase (MGMT) promotor methylation status30,31, Horvath’s methylation age32 and chromosomal copy number analysis were performed as previously described22,33. The DNA methylation-based stemness index, a predictor for oncogenic dedifferentiation, was retrieved according to the stemness index workflow from Malta et al.34 which scales from 0 (low stemness) to 1 (high stemness). We additionally included methylation array data from our previous study22 (GEO with accession GSE116298), which consisted of 58 spatially separated biopsies from 15 adult patients with a pathological diagnosis of glioblastoma IDH-wildtype/IDH-mutant, WHO (2016) grade IV and three patients diagnosed with low-grade meningioma, WHO (2016) grade I. Methylation-based diagnostic classification of the tumor biopsies was performed with the Molecular-neuropathology brain classifier version 12.5 (MNP, https://www.molecularneuropathology.org/mnp). The tumors were examined and stratified into methylation families and methylation family members (subtypes/subclasses) as indicated35,36,37 (Supplementary Table 1).
2021 WHO tumor reclassification
We reclassified the 2016 WHO diagnosis of the patients according to the 2021 WHO classification criteria of CNS tumors20 that relied on tissue-based histological data and molecular information generated at the Sahlgrenska University hospital at the time of diagnosis, and in our laboratory facility at the Sahlgrenska Center for Cancer Research. Molecular data included information on IDH mutation, 1p/19q codeletion, CDKN2A/B homozygous deletion, EGFR amplification, chromosome 7 gain and chromosome 10 loss. The diagnosis of astrocytoma IDH-wildtype, WHO (2016) grade III in patient GU-LGG-93 was changed to glioblastoma IDH-wildtype, CNS WHO grade 4 due to the presence of EGFR amplification and/or chromosome 7 and 10 alterations in the tumor. In the two glioblastomas, IDH-mutant, WHO (2016) grade 4 (GU-HGG-216 and GU-HGG-365), the diagnoses were updated to the newly recognized tumor type astrocytoma IDH-mutant, CNS WHO grade 4. In two other cases (GU-LGG-90 and GU-hgMNG-14), distinct regions of the tumors were detected, corresponding to different 2021 CNS WHO grades and therefore the overall tumor grade was based on the highest grade component present in the tumor, i.e., CNS WHO grade 4 and CNS WHO grade 2, respectively. Molecular reclassification of the diagnoses according to the 2021 WHO CNS criteria is presented in Supplementary Table 2, and it was used further for all analyses herein.
Histology
Fresh-frozen tumor biopsies were fixated in 4% paraformaldehyde (Histolab, Sweden) overnight at 4 °C followed by dehydration in ethanol baths for 12 h and subsequently embedding in paraffin wax. 5 um of formalin-fixed and paraffin-embedded (FFPE) tumor sections were cut using a microtome (HM355S, Thermo Fisher Scientific, Waltham, MA, USA) and stained with Mayer’s hematoxylin and eosin (H&E, Histolab) for tumor purity estimations.
Immunohistochemical staining of IDH1 and MDM2
Immunohistochemical staining of IDH1 and MDM2 was carried out on 5 um FFPE tumor tissue sections. Visualization of neoplastic cells in IDH-mutant gliomas was performed by immunostaining of IDH1 point mutation (R132H) on the DAKO Autostainer 48 Link and Envision FLEX detection system (Dako, Santa Clara, CA, USA). The tumor sections were dried in a heating cabinet for 1 h prior to heat-induced antigen retrieval in high pH-buffer in PT-Link (Dako). The samples were then incubated with a mouse monoclonal IDH1 antibody (DIA-H09, Dianova, Hamburg, Germany) followed by blocking with hydrogen peroxide. Horseradish peroxidase-linked secondary antibodies were added and the staining was visualized using 3,3’-Diaminobenzidine (DAB) chromogen. The sections were counterstained with Mayer’s hematoxylin.
To evaluate MDM2 amplification in meningioma biopsies, the tumor sections underwent heat-induced antigen retrieval with citrate buffer (pH 6.0, Vector Laboratories, Burlingame, CA, USA) followed by blocking of endogenous peroxidase with 3% hydrogen peroxidase in methanol. The samples were stained with the Vectastain ABC kit (Vector Laboratories) using a mouse monoclonal MDM2 antibody (IF2; 1:800, Thermo Fisher Scientific) overnight at 4 °C according to the suppliers instructions with the addition of DAB for detection of antibody signal. The sections were then counterstained with Mayer’s hematoxylin.
Estimation of tumor cell content
Determination of tumor purity was assessed blinded by a specialist in clinical neuropathology with FFPE H&E stained tissue sections from the fresh-frozen tumor biopsies collected in this study and from our previous study22. We estimated tumor purity by evaluating the fraction of neoplastic cells relative to other, nonneoplastic tissue elements such as blood vessels (white blood cells and endothelial cells), immune cells, nonneoplastic glial cells and nerve cells present in the tumor biopsies. The nonneoplastic tissue elements in the samples were considered as factors decreasing the purity of the tumors. Histopathological evaluation of tumor cell content was conducted for 123 out of 126 biopsies since there was insufficient tumor material for histology in one IDH-mutant glioma, CNS WHO grade 2 and two glioblastoma IDH-wildtype, CNS WHO grade 4 biopsies. In three other tumor biopsies (two IDH-mutant gliomas, CNS grade 3 and one glioblastoma IDH-wildtype, CNS WHO grade 4), no neoplastic cells were located by histopathological evaluation and tumor purity was set to 0%. Representative examples of estimation of tumor cell content by H&E stains are shown in Supplementary Fig. 1. In IDH-mutant gliomas, we further examined the reliability of histopathological tumor purity estimates by comparing these with the percentage IDH-mutant positive cells in the tumors determined by immunohistochemistry (Supplementary Fig. 1C, D). Tumor purity was also predicted based on the DNA methylation array data using the publicly available R packages InfiniumPurify25,26, PAMES27, and RF_purify28 (using the RF_absolute and RF_estimate algorithms) as described by the developers. We used methylation array data from 61 adult brain tissue samples17 (GSE109381) as control data set for PAMES and InfiniumPurify when estimating tumor purity in meningioma.
Statistical analyses
All statistical analyses were conducted in R with Rstudio (version 4.0.2). Overall survival (OS) was defined as the time of diagnosis to time of death or last follow-up (June, 17th 2021). The association between OS and stemness index was predicted with the R package Survival38 using the Cox proportional hazards regression model test. Correlation analyses were performed using the R package corrplot39 and Hmisc40 with Pearson correlation. A correlation p value < 0.05 was considered statistically significant. We performed two-tailed t-test to compute the significant difference between tumor groups (p value < 0.05).
Results
Estimation of tumor purity in brain tumors
We sourced between three to five spatially separated tumor biopsies per patient diagnosed with diffuse gliomas, WHO (2016) grade II–IV or meningiomas, WHO (2016) grade II. In total, we collected 68 tumor biopsies from 21 patients and the 2016 WHO diagnoses were updated according to the 2021 WHO classification system, which resulted in 10 IDH-mutant gliomas (four astrocytomas, CNS WHO grade 2–4 and six 1p/19q codeleted oligodendrogliomas, CNS WHO grade 2–3), two glioblastomas IDH-wildtype, CNS WHO grade 4 and nine meningiomas, CNS WHO grade 2 (Supplementary Table 2). We additionally included and reclassified 58 spatially separated biopsies from our previous study22 for further characterization of the intratumor heterogeneity in these tumors, which included one IDH-mutant astrocytoma, CNS WHO grade 4, 14 glioblastomas IDH-wildtype, CNS WHO grade 4 and three meningiomas, CNS WHO grade 1.
We first evaluated the performance of five methods for estimation of tumor purity. Tumor purity was assessed based on pathological evaluation of H&E stained histological samples and calculated with InfiniumPurify, PAMES, RF_absolute and RF_estimate using methylation array data from the 126 biopsies. Tumor purity distributions for each brain tumor type is shown in Fig. 1 and Supplementary Fig. 2. As expected, due to the non-infiltrative and compact growth of meningioma, meningioma biopsies showed higher, and less variable, tumor purity than the diffuse glioma samples. For instance, the mean tumor purity based on histology in meningioma was 84% with a variance of 0.02 compared to IDH-mutant gliomas (mean purity 69%, variance 0.1, p = 9.096e−06) and glioblastoma IDH-wildtype (mean purity 76%, variance 0.05, p = 0.00836). RF_estimate predicted high levels of tumor purity in all three tumor types (median purity >80%). RF_absolute, in contrast, predicted lower tumor purities (median purity <70%) than the other methods. RF_absolute showed the highest correlation with RF_estimate (corr. = 0.89, p < 0.05), which is unsurprising given that they are from the same developer, but InfiniumPurify displayed overall significantly better concordance with all the other methods. Furthermore, histopathological assessment of tumor purity displayed good concordance with InfiniumPurify (corr. = 0.55, p < 0.05) and PAMES (corr. = 0.71, p < 0.05).
Distribution of tumor purity across methylation classes
Next, we analyzed the resulting tumor purity estimates across the methylation classes predicted by the MNP brain classifier version 12.535. The majority of the tumor biopsies (94%, 119/126) were classified with brain tumor methylation class families, which included diffuse glioma IDH-mutant (n = 31), glioblastoma IDH-wildtype (n = 45), meningioma (n = 38), low-grade ganglioglial/neuroepithelial tumor (n = 2) and diffuse pediatric-type high-grade glioma, H3-wildtype and IDH-wildtype (n = 3). The remaining tumor biopsies (n = 7) were classified as normal control tissue, an outcome also previously encountered in tumors with low tumor purity33,36. The vast majority of the tumor biopsies classified as IDH-mutant gliomas (94%, 29/31) had a calibrated family score ≥0.90 (max 1.0) but the median tumor purity differed between the methods (Fig. 2). Similarly, glioblastoma IDH-wildtype (89%, 40/45) had a calibrated score ≥0.90 and a slightly wider tumor purity range compared to IDH-mutant gliomas independent of the measurement method. Classification of meningiomas showed 100% (38/38) of the tumor biopsies with a classification score ≥0.90 and displayed less variation between the tumor purity methods. Compared with the methylation-based tumor purity calculations, the histopathological evaluation provided a more profound discrimination of tumor purity between the tumor methylation classes and the control methylation classes (p < 0.05).
All samples were classified by the MNP methylation-based brain classifier (version 12.5)35 and (A) shows the distribution of tumor biopsies by the methylation class families and class calibrated scores. B Tumor purity estimations among methylation classes as estimated by histology, InfiniumPurify25,26, PAMES27, RF_absolute, and RF_estimate28. Histopathology resulted in a more profound discrimination between the tumor methylation classes and control tissue class.
Diffuse gliomas and high-grade meningiomas are characterized by spatial methylation heterogeneity
The variability in cell type content within tumors has been reported as a potential source of tumor heterogeneity23,24 and we therefore accounted for tumor purity when investigating the intratumor heterogeneity in diffuse gliomas and meningiomas. Tumor biopsies with low tumor purity (less than 70% tumor cell content, n = 21) based on histopathological evaluation were excluded (Supplementary Fig. 1). We chose histopathology for tumor purity estimation since it is used in the clinic, and resulted in a more profound tumor purity discrimination between the tumor methylation class families and the control tissue class (Fig. 2). We further excluded patients with less than three intratumor biopsies and biopsies suspected as normal tissue based on flat copy number alteration (CNA) profiles (generated from the methylation array data). Furthermore, two of the three biopsies that could not be analyzed by histology, GU-HGG-204_3 and GU-HGG-224_2, were excluded due to relatively low tumor purity (mean tumor purity 60%) measured by the methylation-based tools. The third biopsy (GU-LGG-88_6) displayed high tumor purity predicted by InfiniumPurify (92%) and PAMES (87%) and was therefore retained, as these methods correlated well with histopathological evaluation (Supplementary Fig. 2).
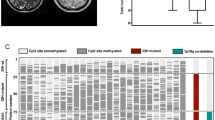
Homogeneous methylation class families and subclasses were found within all IDH-mutant gliomas with the MNP brain classifier (version 12.5)35 (Fig. 3A). Two patients (GU-LGG-73 and GU-LGG-90) showed greater differences in subclass calibrated scores within the tumors (mean difference >30%) compared to the other patients. However, the difference in tumor purity between intratumor biopsies was small (mean difference tumor purity 3%), suggesting that the calibrated scores of the classifier were not influenced by tumor purity in these cases. We previously demonstrated that multiple DNA methylation subtypes coexist within glioblastoma IDH-wildtype, WHO (2016) grade IV, as the tumors (e.g., GU-HGG-271) presented combinations of the RTK1/2 and mesenchymal subtypes22. We found two additional glioblastomas IDH-wildtype, CNS WHO grade 4 (GU-LGG-93 and GU-HGG-287) with subtype heterogeneity, harboring RTK2 and mesenchymal subclasses. The mean tumor purity difference between pairwise intratumor glioblastoma IDH-wildtype biopsies was 6% (range: 3–13%). Differences in tumor purity could thus be ruled out as the confounding factor explaining the subtype heterogeneity in GU-LGG-93 and GU-HGG-271. An overview of glioblastoma IDH-wildtype heterogeneity of representative cases is shown in Supplementary Fig. 3. GU-HGG-271 showed histological appearance of a glioblastoma, CNS WHO grade 4, while GU-LGG-93 showed a less cell-rich tumor. GU-LGG-93 was diagnosed as astrocytoma IDH-wildtype, WHO (2016) grade III but, with additional molecular data generated in this research study (EGFR amplification and copy number variations of chromosomes 7 and/or 10), the tumor was designated as a glioblastoma IDH-wildtype, CNS WHO grade 4 according to WHO 2021 criteria.
A Top panel: Tumor purity estimations based on histopathological evaluation. Each bar represents one biopsy and biopsies from the same patient are grouped together. X denotes a biopsy which could not be analyzed by histology. Middle panel: Biopsy grade based on 2021 WHO criteria. Bottom two panels: Methylation-based class family and subtype/subclass calibrated scores respectively from the MNP methylation-based brain classifier (version 12.5)35. B Intratumor DMPs found in the brain tumors before and after correcting for methylation artifacts of copy number alterations with SeSAMe43. Intratumor DMPs in glioblastoma IDH-wildtype from some samples and the low-grade meningioma (GU-lgMNG) are from Wenger et al.22 GU-LGG lower-grade glioma. GU-HGG high-grade glioma. GU-hgMNG high-grade meningioma. Recurrent tumors are denoted with an R. Patients with subtype/subclass heterogeneity are marked with an asterisk (*) symbol.
Stratification of meningioma by methylation profiling resulted in subtype heterogeneity in three out of eight high-grade meningiomas and one out of three low-grade meningiomas, as these had combinations of the benign, intermediate and/or malignant subtypes. Moreover, two high-grade meningiomas (GU-hgMNG-14 and GU-hgMNG-13R) showed subclass heterogeneity within the intermediate subtype, as the subclasses a and b were present in the tumors. The mean tumor purity difference between pairwise intratumor biopsies in high-grade meningioma was 6% (range: 0–10%), indicating that differences in tumor purity were not causing the observed subtype/subclass heterogeneity in these meningioma patients. For instance, the biopsy with the lowest tumor purity in GU-hgMNG-14R was not the one displaying a different subtype but it was one of the biopsies with as high tumor purity as another intratumor biopsy, i.e., tumor purity was not the underlying factor of the observed subtype heterogeneity. Similarly, the mean tumor purity difference in the low-grade meningioma case with subtype heterogeneity (GU-lgMNG-2) was 3%, which indicated that variation in tumor purity was not causing the observed subtype heterogeneity. An overview of meningioma heterogeneity of representative cases is shown in Supplementary Fig. 4.
Differential methylation at individual CpG sites have been associated with malignant recurrence in glioma41 and meningioma42. We therefore identified and compared the number of differentially methylated CpG sites between intratumor biopsies and examined its association to tumor heterogeneity (Fig. 3B). The mean number of DMPs within IDH-mutant glioma was 16,649 (max: 46,445, min: 3609) and the number of intratumor DMPs were significantly increased in IDH-mutant gliomas of CNS WHO grade 3–4 (mean number of intratumor DMPs: 22,918) compared to CNS WHO grade 2 tumors (mean number of intratumor DMPs: 8289). The mean number of intratumor DMPs in glioblastoma, IDH-wildtype was 21,279 (max: 50,324, min: 820). The mean number of intratumor DMPs was higher in high-grade meningioma (mean: 24,082, max: 68,819, min: 1632) than the previously observed mean of 110 in low-grade meningioma22. This suggests that more malignant tumors are more heterogeneous than tumors of lower-grades. GU-hgMNG-14, which consisted of CNS WHO grade 1 and 2 tumor components, displayed the highest number of intratumor DMPs among meningiomas, likely indicating the underlying tumor biology reflected in the varying tumor grades. We then adjusted for artifactual variation of methylation array data in regions with CNA using the processing pipeline SeSAMe43. The number of intratumor DMPs was slightly reduced; mean number of intratumor DMPs in IDH-mutant glioma: 12,704, glioblastoma IDH-wildtype: 13,599 and high-grade meningioma: 15,393. There was no association between the number of intratumor DMPs and the difference in tumor purity between intratumor biopsy pairs (Supplementary Fig. 5), indicating that intratumor DMPs were not affected by differences in tumor purity.
Spatial heterogeneity in DNA methylation age, stemness index and MGMT promotor methylation
Epigenetic clocks have gained interest for predicting cancer risk and prognosis in adult and pediatric brain tumors and an accelerated epigenetic age has been associated with various methylation-based glioma subtypes32,44,45,46. As previously reported22, we detected variability in methylation age within glioblastoma IDH-wildtype/IDH-mutant, WHO (2016) grade IV, while low-grade meningioma samples exhibited a more homogeneous methylation age. We thus investigated if methylation age differed within IDH-mutant gliomas and high-grade meningiomas using the Horvath clock. IDH-mutant gliomas showed an accelerated methylation age compared to the patients’ chronological age, and methylation age varied within these tumors (mean difference methylation age: 15 years; Fig. 4A). Methylation age was also accelerated in high-grade meningiomas and methylation age differences within these tumors were higher than the previously observed in low-grade meningiomas (mean difference methylation age: 8 and 3 years respectively).
A Methylation age varies within IDH-mutant glioma, glioblastoma IDH-wildtype and high-grade meningioma. The dashed line represents y = x as a reference. B Stemness index differs more in glioblastoma IDH-wildtype and high-grade meningioma compared to IDH-mutant glioma. Methylation age in glioblastoma IDH-wildtype from some samples and low-grade meningioma (GU-lgMNG) are from Wenger et al.22 GU-LGG lower-grade glioma. GU-HGG high-grade glioma. GU-hgMNG high-grade meningioma. Recurrent tumors are denoted with an R. Each biopsy is represented by a circle and color coded according to patient identity.
Recently, the methylation-based stemness index has been proposed as a marker of cancer progression in brain tumors including glioma34. We therefore examined whether there are differences in stemness index in diffuse gliomas and meningiomas, and further analyzed if stemness index correlated with prognosis in glioblastoma IDH-wildtype. We detected a significantly higher stemness index in glioblastoma IDH-wildtype compared to IDH-mutant glioma (mean: 0.25 and 0.10, respectively; p = 6.72e−16; Fig. 4B). Higher differences in stemness index were also detected within glioblastoma IDH-wildtype compared to IDH-mutant glioma (mean: 0.024 vs. 0.012, p = 0.056). We observed a strong correlation between stemness index and the patients’ chronological age in glioblastoma IDH-wildtype (Pearson’s correlation 0.67, p = 5.697e−05) and a negative association to overall survival (p = 3.650e−08). The stemness index in meningioma was also significantly higher in high-grade meningioma compared to low-grade meningioma (mean 0.15 vs. 0.10, respectively, p = 2.64e−05) and it varied intratumorally in one high-grade meningioma patient.
The methylation status of the MGMT promotor is a clinical predictive and prognostic biomarker in glioblastoma47,48 and has also been suggested as a prognostic biomarker in diffuse lower-grade gliomas49. Moreover, we previously found heterogeneous MGMT methylation in the only glioblastoma IDH-mutant, WHO (2016) grade IV patient in a cohort of 12 glioblastomas, WHO (2016) grade IV22. In this study, we therefore assessed MGMT methylation in IDH-mutant gliomas. MGMT promotor methylation status was homogeneous in IDH-mutant glioma CNS WHO grade 2–3 (data not shown).
Heterogeneous copy number alterations within diffuse gliomas and high-grade meningiomas
Heterogeneous chromosomal CNA are found within tumors including glioblastoma9 and meningioma7. To explore the spatial genomic heterogeneity in our cohort of IDH-mutant gliomas and high-grade meningiomas, we derived CNA profiles from the methylation array data for each tumor biopsy. We detected intratumoral differences in copy number aberrations in four out of six analyzed IDH-mutant gliomas. In two of the patients, copy number heterogeneity in chromosome 14q was detected. Homozygous deletion of CDKN2A/B, which has recently been established as a biomarker of grading and prognosis in 2021 WHO IDH-mutant astrocytic gliomas20, was heterogeneous in one out of two IDH-mutant astrocytomas, CNS WHO grade 4 (GU-LGG-90), as two of the three biopsies presented homozygous deletion of CDKN2A/B (Fig. 5A). We did not detect differences in histopathological features within the tumor (Fig. 5B) but, the loss of CDKN2A/B in certain regions of the tumor could likely explain the rapid progression of the patient with a clinical diagnosis of diffuse astrocytoma IDH-mutant, WHO (2016) grade II. Other heterogeneous chromosomal aberrations in GU-LGG-90 included gain of chromosome 1q and focal deletion in chromosome 2q. Furthermore, we observed differences in CNA within three out of eight high-grade meningiomas, where the most common differences were partial or whole loss of chromosomes 4 and 6. Notably, one of the patients (GU-hgMNG-14) showed heterogeneous amplification of the proto-oncogene MDM2. The occurrence of MDM2 amplification is a rare genomic alteration in atypical meningiomas associated with the malignant pathogenesis of the tumor50,51. In this case, the high-level amplification of MDM2 that was found in one out of the three primary biopsies (GU-hgMNG-14_4, Fig. 6A) was also found in all the three recurrent biopsies (GU-hgMNG-14R) (Fig. 6B). Further histopathological examination of the primary tumor in GU-hgMNG-14 revealed a region of the tumor with increased cellularity and presence of larger nucleoli, which might indicate a more aggressive meningioma (GU-hgMNG-14_4) and other regions with histopathological benign phenotypes (GU-hgMNG-14_2 and 3), reflecting the observed CNA heterogeneity in the primary tumor (Supplementary Fig. 4).
A Left panels: Heterogeneous status of MDM2 amplification was identified in the primary tumor of one out of seven high-grade meningioma patients (GU-hgMNG-14). Right panels: MDM2 status was confirmed by immunohistochemical staining of MDM2. B Left panels: Amplification of MDM2 was identified in all biopsies of the recurrent tumor (GU-hgMNG-14R). Right panels: immunohistochemical staining of MDM2. Recurrent tumors are denoted with an R. Scale bars: 50 µm.
Discussion
DNA methylation profiling has gained increasing importance in the decision-making process for tumor diagnostics. Recently, the current 2021 WHO classification system for CNS tumors introduced methylation profiling as an essential diagnostic criterion for certain tumor types, or desirable in many cases, alongside with standard methods20. Heterogeneity in DNA methylation or in chromosomal CNAs, inferred from the methylation array, could thus potentially affect specific biomarkers. Therefore, we aimed to investigate the intratumor genetic and epigenetic heterogeneity in diffuse gliomas and meningiomas through analysis of 126 spatially separated tumor biopsies from 39 adult patients. We evaluated the impact of intratumor variability on methylation-based classification, CNAs and methylation-based biomarkers and investigated tumor purity as a confounding factor in tumor heterogeneity analyses.
We detected intratumor glioblastoma IDH-wildtype, CNS WHO grade 4 heterogeneity in the methylation subclasses, which is in agreement with our previous study22. This was also further supported by a previous study52, where subtype heterogeneity was found in five out of 11 glioblastomas compared to three out of 10 tumors in our study. This difference in frequency can be attributed to the distinct methodologies chosen to approach tumor purity. Intratumor subtype heterogeneity was previously detected in three out of 16 IDH-mutant gliomas52, however we found homogeneity in the methylation class families and subclasses in all IDH-mutant glioma analyzed. Molecular stratification of meningiomas based on DNA methylation profiles has been shown to provide a more accurate prediction of clinical behavior than the WHO classification13,18,19. Intratumor heterogeneity was observed in DNA methylation profiles of high-grade meningiomas6, and we detected multiple methylation subtypes and subclasses coexisting within high-grade meningiomas. In two of these tumors, we observed combinations of the malignant/intermediate subtypes, which could be reflected by the similar biology of these subgroups as previously identified18.
Prediction of age by methylation profiling has been proposed as a prognostic biomarker in diffuse gliomas45,46. We previously demonstrated methylation age heterogeneity within glioblastoma, IDH-wildtype WHO (2016) grade IV and homogeneity in low-grade meningioma22. We therefore calculated methylation age in the present study and found it to be heterogeneous within IDH-mutant gliomas and high-grade meningiomas. Similarly, the predicted stemness index significantly increased with tumor grade in our cohort, which is in agreement with Malta et al.34. Furthermore, we found greater variability in the stemness index between and within glioblastoma IDH-wildtype compared to IDH-mutant glioma, and also between high-grade meningioma compared to low-grade meningioma. Further validation with larger cohorts is therefore required to validate the prognostic value of these biomarkers.
The 2021 WHO criteria has recognized CDKN2A/B homozygous deletion as a biomarker of grading and prognosis in IDH-mutant astrocytic gliomas and the presence of CDKN2A/B homozygous deletion designates a CNS WHO grade 4 tumor20. We identified intratumor heterogeneity of the CDKN2A/B deletion in one of the two IDH-mutant astrocytomas, CNS WHO grade 4 in our cohort after excluding samples with low tumor purity. To the best of our knowledge, intratumor heterogeneity of CDKN2A/B has previously not been reported. Given that this alteration is a late event associated with malignant transformation it could potentially explain this finding. Variability in CDKN2A/B copy number alteration indicates that there is a risk of sampling bias that may lead to misclassification of tumor grade in IDH-mutant astrocytomas. This in turn may have implications on the therapeutic approach as according to guidelines, a wait-and-scan procedure is performed following a (near) radical surgery of diffuse lower-grade gliomas IDH-mutant3. Intratumor copy number heterogeneity was also observed in one high-grade meningioma affecting the MDM2 proto-oncogene. Amplification of the MDM2 is a rare genomic alteration in high-grade meningioma and it is considered to drive malignant progression50,51. In our study, the presence of MDM2 amplification varied within the primary tumor in patient GU-hgMNG-14, but it was stable in the recurrent tumor, thus reflecting the malignant progression of the tumor. Intratumor heterogeneity of MDM2 could potentially have implications when evaluating the clinical outcome of meningioma patients, for instance in clinical trials of targeted therapeutics with MDM2 inhibitors. Further studies are needed to improve our understanding of MDM2 heterogeneity and its clinical impact in meningioma patients.
Methylation values are based on the type of cells constituting the tumor tissue, including cancer cells and normal cells such as immune cells, which could contribute to tumor heterogeneity. We therefore evaluated the performance of different methods to assess tumor purity in our cohort: histopatholology, InfiniumPurify, PAMES, RF_absolute and RF_estimate. Histopathological evaluation was in good agreement with the methylation tools InfiniumPurify and PAMES and could better discriminate between tumor methylation classes from the normal tissue class than the other methods. One should note that the different computational methods are based on the same material, whereas histology was made on an adjacent tumor section from the same biopsy. In agreement with Johann et al.28 RF_estimate systematically predicted higher tumor purities in diffuse gliomas than the other methods and RF_absolute estimated the lowest tumor purities independently of the tumor type. This warrants some cautions when setting a general threshold when selecting samples for further analysis as it may vary depending on the method, the type of tumor, as well as the specific research question. We included biopsies with >70% tumor purity, as previously recommended17,36, for intratumor heterogeneity analyses. The intratumor subtype heterogeneity found in glioblastoma IDH-wildtype and high-grade meningioma was not driven by differing tumor content within the tumors in most of the cases. The intratumor heterogeneity in diffuse glioma and meningioma was also detected at individual CpG sites. We observed that the number of DMPs was not associated with differences in tumor purity between intratumor biopsy pairs demonstrating that variable tumor content was not causing such methylation heterogeneity in these tumors. The high intratumor DMP heterogeneity found in patient GU-hgMNG-14 could, however, be explained by the distinct phenotypes with clearly two tumor components and corresponding different CNS WHO grades of these.
In conclusion, our study demonstrates the variability in DNA methylation within diffuse gliomas and meningiomas that potentially affects tumor classification and interpretation of methylation-based biomarkers. Using chromosomal copy number profiles inferred from the array data, we also demonstrate CNA heterogeneity, with implications for clinical diagnosis and prognosis.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ostrom Q, Patil N, Cioffi G, Waite K, Kruchko C & Barnholtz-Sloan J. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro Oncol. 22, iv1-iv96 (2020).
Goldbrunner R, Stavrinou P, Jenkinson M, Sahm F, Mawrin C, Weber D et al. EANO guideline on the diagnosis and management of meningiomas. Neuro Oncol. 23, 1821–1834 (2021).
Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn J, Minniti G et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 18, 170–186 (2021).
Dagogo-Jack I & Shaw A. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018).
Ceccarelli M, Barthel F, Malta T, Sabedot T, Salama S, Murray B et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 164, 550–563 (2016).
Magill S, Vasudevan H, Seo K, Villanueva-Meyer J, Choudhury A, John Liu S et al. Multiplatform genomic profiling and magnetic resonance imaging identify mechanisms underlying intratumor heterogeneity in meningioma. Nat. Commun. 11, 4803 (2020).
Pfisterer W, Hank N, Preul M, Hendricks W, Pueschel J, Coons S et al. Diagnostic and prognostic significance of genetic regional heterogeneity in meningiomas. Neuro Oncol. 6, 290–299 (2004).
Sottoriva A, Spiteri I, Piccirillo S, Touloumis A, Collins V, Marioni J et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A. 110, 4009–4014 (2013).
Sturm D, Witt H, Hovestadt V, Khuong-Quang D, Jones D, Konermann C et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012).
Network CGAR. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New Engl. J. Med. 372, 2481–2498 (2015).
Verhaak R, Hoadley K, Purdom E, Wang V, Qi Y, Wilkerson M et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010).
Yan H, Parsons D, Jin G, McLendon R, Rasheed B, Yuan W et al. IDH1 and IDH2 mutations in gliomas. New Engl. J. Med. 360, 765–773 (2009).
Nassiri F, Liu J, Patil V, Mamatjan Y, Wang J, Hugh-White R et al. A clinically applicable integrative molecular classification of meningiomas. Nature 597, 119–125 (2021).
Jones P, Issa J & Baylin S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 17, 630–641 (2016).
Baylin SB & Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 8 (2016).
Danielsson A, Nemes S, Tisell M, Lannering B, Nordborg C, Sabel M et al. MethPed: a DNA methylation classifier tool for the identification of pediatric brain tumor subtypes. Clin. Epigenetics, 2015, Vol. 7 7 (2015).
Capper D, Jones D, Sill M, Hovestadt V, Schrimpf D, Sturm D et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018).
Sahm F, Schrimpf D, Stichel D, Jones D, Hielscher T, Schefzyk S et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 18, 682–694 (2017).
Olar A, Wani K, Wilson C, Zadeh G, DeMonte F, Jones D et al. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 133, 431–444 (2017).
Louis D, Perry A, Wesseling P, Brat D, Cree I, Figarella-Branger D et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 23, 1231–1251 (2021).
Jackson RJ, Fuller GN, Abi-Said D, Lang FF, Gokaslan ZL, Shi WM et al. Limitations of stereotactic biopsy in the initial management of gliomas. Neuro Oncol. 3, 193–200 (2001).
Wenger A, Ferreyra Vega S, Kling T, Olsson Bontell T, Jakola AS & Carén H. Intratumor DNA methylation heterogeneity in glioblastoma: implications for DNA methylation-based classification. Neuro Oncol. 21, 616–627 (2019).
Jaffe A & Irizarry R. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31 (2014).
Aran D, Sirota M & Butte A. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015).
Zheng X, Zhang N, Wu H & Wu H. Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies. Genome Biol. 18, 17 (2017).
Qin Y, Feng H, Chen M, Wu H & Zheng X. InfiniumPurify: An R package for estimating and accounting for tumor purity in cancer methylation research. Genes & diseases 5, 43–45 (2018).
Benelli M, Romagnoli D & Demichelis F. Tumor purity quantification by clonal DNA methylation signatures. Bioinformatics 34, 1642–1649 (2018).
Johann P, Jager N, Pfister S & Sill M. RF_Purify: a novel tool for comprehensive analysis of tumor-purity in methylation array data based on random forest regression. BMC Bioinformatics 20, 428 (2019).
Zheng X, Zhao Q, Wu H, Li W, Wang H, Meyer C et al. MethylPurify: tumor purity deconvolution and differential methylation detection from single tumor DNA methylomes. Genome Biol. 15, 419 (2014).
Bady P, Delorenzi M & Hegi ME. Sensitivity Analysis of the MGMT-STP27 Model and Impact of Genetic and Epigenetic Context to Predict the MGMT Methylation Status in Gliomas and Other Tumors. J. Mol. Diagn. 18, 350–361 (2016).
Bady P, Sciuscio D, Diserens A-C, Bloch J, den Bent M, Marosi C et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 124, 547–560 (2012).
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
Ferreyra Vega S, Olsson Bontell T, Corell A, Smits A, Jakola AS & Carén H. DNA methylation profiling for molecular classification of adult diffuse lower-grade gliomas. Clin. Epigenetics 13, 102 (2021).
Malta T, Sokolov A, Gentles A, Burzykowski T, Poisson L, Weinstein J et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 173, 338–354 e315 (2018).
Molecular neuropathology. Brain classifier 12.5, https://www.molecularneuropathology.org/mnp (2022).
Capper D, Stichel D, Sahm F, Jones D, Schrimpf D, Sill M et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 136, 181–210 (2018).
Maas S, Stichel D, Hielscher T, Sievers P, Berghoff A, Schrimpf D et al. Integrated Molecular-Morphologic Meningioma Classification: A Multicenter Retrospective Analysis, Retrospectively and Prospectively Validated. J. Clin. Oncol. 39, 3839–3852 (2021).
Therneau T. A Package for Survival Analysis in R, R package version 3.2–3, https://CRAN.R-project.org/package=survival (2021)
Wei T & Simo V. R package ‘corrplot’: Visualization of a Correlation Matrix, R package version 0.84, https://github.com/taiyun/corrplot (2021)
Harrell JF. Harrell Miscellaneous, R package version 4.4–1, https://CRAN.R-project.org/package=Hmisc (2021)
de Souza C, Sabedot T, Malta T, Stetson L, Morozova O, Sokolov A et al. A Distinct DNA Methylation Shift in a Subset of Glioma CpG Island Methylator Phenotypes during Tumor Recurrence. Cell Rep. 23, 637–651 (2018).
Millesi M, Ryba AS, Hainfellner JA, Roetzer T, Berghoff AS, Preusser M. et al. DNA Methylation associates with clinical courses of atypical meningiomas: A matched case-control study. Front. Oncol., 603, 811729, (2022).
Zhou W, Triche T, Jr., Laird P & Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 46, e123 (2018).
Kling T, Wenger A & Carén H. DNA methylation-based age estimation in pediatric healthy tissues and brain tumors. Aging (Albany N. Y.) 12, 21037–21056 (2020).
Liao P, Ostrom Q, Stetson L & Barnholtz-Sloan J. Models of epigenetic age capture patterns of DNA methylation in glioma associated with molecular subtype, survival, and recurrence. Neuro Oncol. 20, 942–953 (2018).
Zheng S, Widschwendter M & Teschendorff A. Epigenetic drift, epigenetic clocks and cancer risk. Epigenomics 8, 705–719 (2016).
Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Eng. J. Med. 352, 997 (2005).
Malmstrom A, Gronberg B, Marosi C, Stupp R, Frappaz D, Schultz H et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 13, 916–926 (2012).
Bell E, Zhang P, Fisher B, Macdonald D, McElroy J, Lesser G et al. Association of MGMT Promoter Methylation Status With Survival Outcomes in Patients With High-Risk Glioma Treated With Radiotherapy and Temozolomide An Analysis From the NRG Oncology/RTOG 0424 Trial. Jama Oncology 4, 1405–1409 (2018).
Weber R, Bostrom J, Wolter M, Baudis M, Collins V, Reifenberger G et al. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc. Natl. Acad. Sci. U. S. A. 94, 14719–14724 (1997).
Wylleman R, Debiec-Rychter M & Sciot R. A rare case of atypical/anaplastic meningioma with MDM2 amplification. Rare Tumors 10, 2036361318779511 (2018).
Verburg N, Barthel F, Anderson K, Johnson K, Koopman T, Yaqub M et al. Spatial concordance of DNA methylation classification in diffuse glioma. Neuro Oncol. 23, 2054–2065 (2021).
Acknowledgements
We thank UCL Genomics for EPIC array processing.
Funding
This work was supported by the Swedish Cancer Society, the Swedish Research Council and the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement, the Swedish innovation agency Vinnova and the research foundations of Assar Gabrielsson and Wilhelm & Martina Lundgren. Open access funding provided by University of Gothenburg.
Author information
Authors and Affiliations
Contributions
SFV, AW, TK, and HC designed the study and HC coordinated it. ASJ conducted the surgical procedures and provided clinical data and input to the study. SFV performed all experimental procedures with the exception of the immunohistochemical and histopathological evaluations, which were performed by TOB. SFV performed the data analysis and prepared the figures and tables with assistance from all authors. All authors read and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the regional ethics committee in the Västra Götaland region in Sweden (Dnr 604–12 and Dnr 1067-16). The study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ferreyra Vega, S., Wenger, A., Kling, T. et al. Spatial heterogeneity in DNA methylation and chromosomal alterations in diffuse gliomas and meningiomas. Mod Pathol 35, 1551–1561 (2022). https://doi.org/10.1038/s41379-022-01113-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-022-01113-8
This article is cited by
-
Longitudinal DNA methylation analysis of adult-type IDH-mutant gliomas
Acta Neuropathologica Communications (2023)
-
Utility of genome-wide DNA methylation profiling for pediatric-type diffuse gliomas
Brain Tumor Pathology (2023)
-
DNA methylation alterations across time and space in paediatric brain tumours
Acta Neuropathologica Communications (2022)
-
Regulatory function of DNA methylation mediated lncRNAs in gastric cancer
Cancer Cell International (2022)