Abstract
Metaplastic breast carcinomas are a rare and heterogeneous group of tumors (0.5–2%). They are mainly triple negative tumors but they present poorer chemotherapy responses and worse prognosis than other triple negative tumors. The aim of our study was to characterize the molecular profile and tumor evolution in matched (primary-relapse) tumor samples from patients with early-stage metaplastic breast carcinomas who had disease recurrence/progression. We performed genomic profiling of tumor biopsies at least from two different time points of their tumor evolution. Tumor samples were analyzed by DNA-Next Generation Sequencing (Illumina 2 x 75bp) using the Action OncoKitDX panel (Imegen-Health in Code group), which includes point mutations in 50 genes, CNVs, and fusion genes. Only pathogenic and likely pathogenic variants were considered for analysis and they were categorized following the ComPerMed criteria. We analyzed 21 matched tumor samples (8 primary and 13 relapse/progression samples). Genomic profiling of matched tumor samples revealed that mutations present in primary tumors are generally maintained in the relapse/disease progression. We did not find a significant increase in point mutations between primary and relapse/progression samples, although gene amplifications were found more frequently in relapse/progression samples. Tumor samples harbored high frequency of TP53 (100%) and TERT promoter (29%) mutations, and of MYC amplifications (80% of which in relapse/progression samples). No PI3KCA mutations were found, but PTEN variations were enriched in 38% of samples (10% mutations and 28% deletions). FGFR1 amplifications were identified in 13% of samples (primary tumor only). Neither ERBB2 nor EGFR gene amplifications were detected. The most frequent pathogenic alterations occurred in cycle regulation’s genes, including TP53 and TERT promoter mutations, and MYC amplifications. Relapse/progression samples were highly enriched for MYC amplification. Larger studies are required to better characterize these tumors, and identify new strategies to improve the prognosis of these patients.
Similar content being viewed by others
Introduction
Metaplastic breast carcinoma is a heterogeneous and rare subtype that constitutes 0.25 to 2% of all breast cancers1,2. These tumors are histologically defined by differentiation of neoplastic epithelium into squamous or mesenchymal-like elements.
Clinicopathologic features of metaplastic breast carcinomas include larger size, higher histological grade, less lymph node involvement, and less vascular invasion than invasive carcinoma of no special type (ductal)3,4. Most metaplastic breast carcinomas (>90%) lack the expression of estrogen and progesterone receptors and HER2, which leads to their classification as triple negative breast cancers (TNBC). The 5th edition of WHO classification of breast tumors categorizes metaplastic breast carcinomas as mixed metaplastic carcinoma, low-grade adenosquamous carcinoma, fibromatosis-like metaplastic carcinoma, squamous cell carcinoma, spindle cell carcinoma, and metaplastic carcinoma with heterologous mesenchymal differentiation5,6. Unlike TNBC of no special type, metaplastic carcinomas are less responsive to standard chemotherapy treatments, and are associated with worse survival outcomes7,8.
Some studies have suggested that metaplastic breast carcinomas may derive from undifferentiated pluripotent stem-cell-like cells9. Molecular analysis has shown that metaplastic breast carcinomas are enriched in stem cell and epithelial-mesenchymal transition (EMT) features, which has led to their classification within the claudin-low subgroup. This subgroup is characterized by being enriched in EMT, immune response and stem-cell process markers10,11,12 involved in drug resistance, increased invasiveness, and the development of metastases. Furthermore, the co-occurrence of an EMT phenotype and alterations in PI3KCA would lead to increased aggressiveness of these tumors10. There are no specific pathognomonic mutations identified for metaplastic breast carcinomas. In several studies comparing metaplastic breast carcinomas to other TNBC, PI3K/AKT/mTOR,Wnt/β-catenin signaling pathway alterations, MYC and TP53 alterations and EGFR amplifications were the most frequently detected variants10,13,14. Nonetheless, all of these studies were retrospective, based on small and heterogeneous populations with no paired tumor samples, and were carried out using different molecular techniques.
The aim of our study was to characterize the genomics and tumor evolution in matched (primary-relapse) samples of patients with metaplastic breast cancer.
Materials and methods
Study design and patient population
A search for patients with metaplastic breast carcinoma treated between 2009 and 2020 was conducted at Catalan Institute of Oncology-University Hospital of Bellvitge, L’Hospitalet (Barcelona). Only patients with at least two tumor samples available in the Pathology department (one of the primary tumor and other of the recurrence/progression) were selected. Data regarding histologic type, hormone receptor and HER2 status were collected from pathologic reports. Tumor stage and other clinical data, including treatment information were obtained from the electronic medical record. Study protocol was approved by the institutional review board at University Hospital of Bellvitge-IDIBELL (BB20-017). Informed written consent was obtained from all patients alive at the moment of the analysis.
Tumor samples
All samples of metaplastic breast carcinomas were provided by Biobank HUB-ICO-IDIBELL, integrated in the Spanish Biobank Network and funded by Instituto de Salud Carlos III (PT17/0015/0024) and by Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d’Oncologia de Catalunya (XBTC). Tumor samples were reviewed by breast pathologists (TS, JB, and AP) and classified according to the latest WHO classification of Breast tumors15. Representative sections of formalin-fixed paraffin-embedded blocks of each metaplastic breast carcinoma were selected and used for DNA extraction and sequenced through Action OncoKitDx NGS gene panel (Imegen-Health in Code Group). A total of 21 paired samples from 8 patients (P1-P8) were included in the study. Gene panel sequencing was performed in 8 primary tumor samples, and their respective 13 recurrences (loco-regional or metastases).
Genomic DNA (gDNA) from the included samples was extracted with the RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE (Thermo Fisher Scientific) and the QIAamp DNA Investigator Kit (Qiagen). DNA concentration was quantified with the Qubit Fluorimeter (Thermo Fisher Scientific) with Qubit dsDNA BR Assay kit and Qubit dsDNA HS Assay kit (Invitrogen). The DNA Integrity Number (DIN) of the different samples was determined using the DNA ScreenTape assay (Agilent Technologies). Cases with a DIN value equal to or higher than 2.4 were considered despite the recommended cut-off DIN value of 3 by the Action OncoKitDx user guide. This cut-off value does not imply that there are no guarantees of a reliable sequencing result. The decision of including samples under DIN of 3 was made considering the limited number of samples available in the study. The samples finally included in the study obtained correct quality parameters in all sequencing steps.
NGS sequencing and bioinformatics pipeline
Tumor samples were analyzed by DNA Next Generation Sequencing (Illumina 2 x 75bp) using the Action OncoKitDX panel which studies point mutations in 50 genes, copy number variants (CNVs) throughout the genome, and fusion genes among 8 genes with any other partner of the genome. It also determined Microsatellite Instability (MSI) through 110 markers and identified pharmacogenetic SNPs associated with treatment toxicity or efficacy according to PharmGKB. All tumor suppressors or oncogenes currently within the standard of care of a high number of adult solid tumors were covered (Supplementary material 1). The results were classified following the recommendations of the American College of Medical Genetics (ACMG)15.
Bar-coded libraries were amplified and sequenced on the NextSeq 550 system (Illumina) for massive library sequencing in “Stand-alone” mode with 2 × 75 paired-end reads following the manufacturer’s instructions. The Sample Sheet was generated using the Illumina Experiment Manager (IEM) software version 1.14.0 (Illumina). The monitorization of sequencing run quality was based on the Q30 value and cluster pass filter, setting a threshold at 80% and 70% respectively. The FASTQ files generated followed a quality evaluation applying the FastQC v0.11.5 software (Babraham Bioinformatics). QC metrics were also evaluated from the BAM file of each sample trough uniformity, average coverage and percentage of the region covered at 100x. Bioinformatic analysis, including the alignment to the reference sequence Genome Reference Consortium Human Build 37 (GRCh37), annotation and variant calling, followed a self-developed pipeline through the DataGenomics platform. The variant assessment and categorization were performed as previously reported by Martínez-Fernández, P16.
Results
Patient’s and tumor characteristics
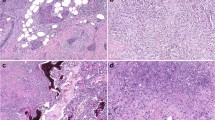
Between January 2009 and December 2020, 73 patients were diagnosed of metaplastic breast carcinoma in our institution, out of a total of 6000 breast cancers diagnosed (1, 2%). Among them, we selected tumor samples from those patients with both primary tumor and relapse/progression tissue available meeting the quality requirements for molecular study. We finally analyzed 21 matched primary-relapse tumor samples from eight patients that met those criteria: 8 primary tumor samples and 13 loco-regional/metastatic samples. In all patients’ genomic characterization was performed at least at two different time points of their tumor evolution, but in three patients, genomic characterization was performed at three different time points, and in one patient at four time points. Baseline patient and tumor characteristics and first site of recurrence are listed in Table 1. The most common histology at diagnosis was spindle cell carcinoma in 3 patients (37%) (Fig. 1A). In two patients although the initial diagnosis was invasive carcinoma of no special type (ductal), the relapsed tumor was a metaplastic carcinoma, so that they were also included in this series. All except one case (in which progesterone receptor was 3%) had a triple negative profile and a high proliferation index measured by ki67 (between 30–80%). In the early setting, all patients received chemotherapy+/− radiotherapy per institutional guidelines. The histologic subtype of primary metaplastic breast carcinoma and their matched metastatic/recurrence were concordant in four cases whereas the morphology changed in four. Moreover in patient 2 (P2) the last recurrence (P2-R3) changed the morphologic subtype from squamous cell carcinoma to spindle cell carcinoma (Fig. 2). The most frequent histology of the first site of recurrence was metaplastic carcinoma with heterologous mesenchymal differentiation (chondroid) (Fig. 1B), found in 5 patients (62%). Tumor evolution and clinical outcome of the eight patients included in this study and pathogenic gene variants identified are summarized in Fig. 3.
Primary metaplastic carcinoma with squamous differentiation (A). Local recurrence of metaplastic squamous cell carcinoma with abundant keratinization and marked nuclear atypia (B). Metastatic metaplastic squamous cell carcinoma in cranial vault infiltrating cerebral tissue (C). Metastatic metaplastic spindle cell carcinoma of the skull featuring high-grade spindled cells and abundant mitoses (D). Scale bars equal 50 μm on (A, B, and D), and 100 μm on (C).
Molecular alterations of metaplastic carcinomas
Tumor genomic characterization detected in each patient based on the different time points of their tumor evolution are listed in Table 2.
Cell cycle regulation
The most frequently mutated gene among both primary tumor and metastases samples identified was TP53. A pathogenic variant within TP53 was identified in all samples (100%) but just 24% of sequenced samples also carried a TP53 mono-allelic deletion (P1, P5, and P6). The allelic frequency of TP53 variants was higher among metastases than primary tumor samples of these three cases (Fig. 3).
Mutations within the TERT promoter were detected in 2 (P3 and P8) out of 8 cases (25%) and they were observed in both primary tumor and metastatic/relapse samples. Both cases were a spindle cell carcinoma subtype at diagnosis but metastatic relapses of P3 changed and were classified as mixed metaplastic carcinoma (spindle cell and mesenchymal chondroid differentiation), keeping though the same mutational pattern.
MYC amplification was observed in 3 cases (P2, P6, and P7) out of 8 cases (37,5%) and in 24% of the 3 samples. Among these samples, 80% were samples from metastases and only 20% from primary tumors. Regarding tumor subtypes, no clear relationship was observed; this alteration was found in mesenchymal chondroid differentiation (P6-R1, P7-PT, and P7-R1), squamous cell carcinoma (P2-R2), and spindle cell carcinoma (P2-R3).
PTEN/PI3K/AKT signaling pathway and MAPK pathway
PTEN alterations were frequently detected in our study of paired samples. Although PTEN mutations were present exclusively in 1 out of 8 patients (13%), a mono-allelic PTEN deletion was detected in 38% of the analyzed samples. Overall, a PTEN gene variation (mutation or deletion) was observed in 41% of the patients (P2, P4, P7, and P8). The samples corresponded to different tumor subtypes: squamous (P2-R2), spindle cell (P2-R3, P4-PT, P4-R1 and P8-R1, P8-R2), and, heterologous mesenchymal differentiation (chondroid) (P7-PT and P7-R1). It should be noted that in P4, this alteration was already present in the primary tumor (invasive carcinoma of no special type). Patient 7 samples showed co-existence of PTEN loss and MYC amplification. A PIK3CA amplification was detected exclusively in one patient (P7 carcinoma with heterologous mesenchymal differentiation, chondroid) and this alteration was identified in both the primary tumor and the relapse samples. None of the tumor samples analyzed harbored mutations in PIK3CA, AKT (AKT1, AKT2, AKT3) nor in the MAPK pathway genes included in the panel (NRAS, HRAS, KRAS, MAP2K1).
Tyrosine kinase receptors EGFR, ERBB2, and FGFR1
Among the sequenced samples, no EGFR or ERBB2 gene amplifications were observed. EGFR gene gain (aneusomy) was observed in 25% of cases (P2, P3). An amplification of FGFR1 was described in the primary tumor of one patient (P8), but it was lost in the metastatic sample.
Other gene alterations
In the tumor samples analyzed, other SNVs and CNVs have been detected in specific genes from particular cases. Among them, a BRCA1 mutation was identified in one out of the eight patients (P6). The pathogenic mutation had a greater allelic frequency in the metastatic samples than in the primary tumor (VAF 3.85% in primary tumor vs 23.10% and 70.45% in paired metastases). No mutation was detected in ARID1A or CTNNB1 genes. A ZNF217 amplification (8–10 copies) was detected in both samples of one patient (P7). Additionally, a focal amplification of more than twenty copies was evidenced throughout 9p23-p22, including NFIB gene. It was observed in one patient, but solely in the most recent metastasis of this patient (P2-R3).
Structural events and Microsatellite instability
The gene panel used studies gene fusions and gains and losses of complete or part of chromosomes; both were reported as structural events. Overall, the sequenced samples were characterized by a high number of chromosomal copy number aberrations. This fact was proved in both primary tumor samples and their corresponding recurrence samples. Each sample carries an average number of eighteen large copy number variants, considering those of at least 6 kb in length (data not shown). No gene fusions of target genes were detected among the sequenced samples. Deletions, gains, and amplifications of single genes with clinical significance are reported in previous sections and complete data about mutations and CNVs to highlight is reported in Table 2.
Microsatellite stability was valued among all the samples. The majority of the analyzed samples (87%) presented values compatible with stable tumors (MSS) and only 13% of the cases presented an MSI consistent with low-grade unstable tumors (MSI-L).
Primary tumor vs recurrences
With regard to single nucleotide variants (SNV), few differences were found in terms of mutation-carrying genes between primary tumor and metastases samples. All genes mutated among metastasis samples were already mutated in their corresponding primary tumor. Overall, TP53 mutations had greater allelic frequency among metastatic samples than among primary tumors. Concerning focal copy number variations (CNVs) involving genes previously implicated in metaplastic breast cancer, some disparities were detected between primary tumors and their corresponding metastases. In addition to an increased recurrence of MYC amplifications among metastatic samples (patients P2, P6, P8) reported above, a BRCA1 deletion was identified exclusively in a metastatic tumor (P6).
Discussion
Metaplastic breast carcinomas are characterized by histological and molecular heterogeneity, as well as poor survival outcomes. However, little is known about their molecular profile and tumor evolution. In the present study, genomic profiling of matched (primary-relapse) tumor biopsies of patients with metaplastic breast carcinoma revealed that mutations present in primary tumors are generally maintained in the relapse/disease progression samples, and only few tumor samples harbored new alterations, such as MYC amplification, which was enriched in the relapse/progression samples.
Importantly, all samples analyzed in this study, either from primary or relapsed paired tumors, carried TP53 mutations. These findings suggest that TP53 mutations play a prominent role in metaplastic breast carcinoma, and that dysregulation of this tumor suppressor gene occurs early during tumor evolution. However, despite the high prevalence of TP53 mutations and the coexistence of TP53 deletions, none TP53 variant presented an allelic frequency compatible with a double hit affecting all tumor region analyzed in this study. In prior studies, TP53 mutations have been identified in up to 70% of patients13,17,18,19,20.
In our series, mutations within the TERT promoter were detected in two patients (6 of out 21 samples: 28%). Interestingly and similar to which occurred with TP53 mutations, TERT promoter mutations were observed in all tumor samples analyzed from these two patients (primary tumors and two additional time points of their tumor evolution). Of note, both cases were spindle cell carcinoma subtype (67% of all spindle tumor cases). In contrast to other types of tumors, TERT promoter mutations have been rarely observed in invasive breast cancers (<1%)21. However, Krings G et al. also described a 25% incidence of TERT alterations in their series of metaplastic breast carcinomas, specifically in tumors with spindle (47%) and/or squamous differentiation, but not matrix-producing carcinomas17. These mutations are also considered to be an earliest genetic event in tumorigenesis22. PIK3CA mutations have been previously described as one of the most prevalent alterations in metaplastic breast carcinomas, being identified in 30–50% of cases10,15,22,23. Data from 19 metaplastic breast carcinoma included in c-BioPortal, showed a 26.3% of PI3KCA mutated samples. Most of the PIK3CA mutations detected were activating, causing deregulation of the pathway, and they have been associated with worse prognosis than non-carriers. However, in our study a PIK3CA amplification was detected only in one patient but none of the tumor samples analyzed harbored mutations in PIK3CA, AKT or in the MAPK pathway, despite an adequate gene coverage by the panel16. This difference in incidence of PI3KCA alterations could possibly be due to the small number of samples in our and others published series, which could lead to sample selection bias. Detecting PIK3CA mutations in metaplastic breast carcinoma may lead to PIK3a inhibitors recommendation within a clinical trial24 although the evidence is still scarce.
MYC is the most frequently amplified gene in metaplastic breast cancer. MYC copy number variations have been reported recurrently among different patient series, being identified in more than 17% of patients14. The role of MYC in metaplastic breast carcinoma is poorly understood and the evolutions of MYC alterations between diagnosis and tumor relapse have not been well defined. We detected MYC amplifications in 24% of the total samples analyzed. Interestingly, most of MYC amplification cases corresponded to relapsed tumor samples (80%). These findings suggest that MYC amplification would not be a recurrent driver at diagnosis but rather an oncogenic promoter during tumor development25. Additionally, MYC amplifications have been described more frequently in squamous or spindle cell differentiation26 however, such morphologic correspondence was not found in our series except for one case with MYC amplification that corresponded to squamous cell carcinoma (P1).
No EGFR gene amplification was observed among the sequenced samples. However, EGFR gene gain (aneusomy) was detected in 25% of cases. These results are in line with previously published data; although it may be over-expressed in one-third of metaplastic breast carcinomas, no activating mutations have been commonly found in EGFR26,27.
NFIB (Nuclear Factor IB; 9p23-p22) amplification was detected in the most recent relapse sample analyzed from patient number 2 (P2). NFIB has been repeatedly amplified or overexpressed in TNBC. This last relapse does not present a squamous differentiation as the previous ones, but acquires spindle cell traits. Therefore, and at least in this patient, NFIB gene amplification is associated with an advanced disease stage and might be related to the histological evolution identified, since it represents the main molecular difference with respect to previous samples.
ZNF217 (Zinc Finger Protein 217) amplification was detected only in patient 7, which falls within a mesenchymal subtype with chondroid differentiation. The involvement of ZNF217 overexpression in the epithelial-mesenchymal transition has been previously described in breast cancer28 and it is associated with poor prognosis29. In fact, it may promote bone metastases in breast cancer30. However, the involvement of ZNF217 in metaplastic breast carcinoma has not been well established. In our series, it is detected in 13% of cases (P7) and it was observed both in the primary and relapsed tumor samples. The patient did not present bone metastatic relapses during the tumor evolution. Despite ZNF217 may be implicated in the tumor development and the possible association of this biomarker with a poor prognosis, more data are required to be able to transfer clinical information in patients with metaplastic breast cancer when ZNF217 amplification is detected. Wnt/β-catenin pathway has been proposed as one of the pathways most implicated in MBC oncogenesis. Different series observed alterations in this pathway in around 50–90% of metaplastic breast carcinomas13,31. Unfortunately, genes involved in this pathway are not the target of the panel, and therefore, we do not have information regarding them.
Relationship between different genetic alterations and histological subtypes is poorly understood and the evolution of these mutations during tumor progression is unknown. Due to the small size of our series, we could not establish any relationship between mutational profile and pathological subtypes of metaplastic breast carcinomas.
While genetic biomarkers are scarce in metaplastic breast cancer, some of the reported alterations may transfer clinical useful information. None diagnostic, prognostic, therapeutic or resistance to therapy tier I variants were identified but several Tier II alterations were detected. More precisely, tier II variants were identified among 63% of patients (38% of total samples). MYC amplification has been reported as a therapy resistance marker25. PTEN loss of function mutations and gene deletions has been described as a marker of resistance to PI3K-p110-alpha selective inhibitors such as alpelisib. Therefore, and although clinical evidence is still lacking, the search for actionable markers in this tumor type seems essential24.
The major limitation of our series is the small number of patients who can carry out unrepresentative results. In addition, it must be taken into account that only patients with accessible disease for biopsy at the time of relapse/progression have been included, which may condition the results.
To our knowledge, this is the first series to compare the molecular profile of matched tumor samples of metaplastic breast carcinomas. These data suggest that mutations present in primary tumors are maintained in disease relapse/progression and only a few new alterations emerged in the tumor evolution of metaplastic breast carcinomas, such as MYC amplification. Due to the small sample size of the series, no relationship between the mutational profile and the pathological subtypes of metaplastic breast carcinomas was established. A better understanding of the pathogenesis of this group of tumors may lead to individualized treatment approaches in order to improve the prognosis of these patients.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Pezzi, C. M. et al. Characteristics and treatment of metaplastic breast cancer: Analysis of 892 cases from the national cancer data base. Ann. Surg. Oncol. 14, 166–173 (2007).
Weigelt, B., Eberle, C., Cowell, C. F., Ng, C. K. Y. & Reis-Filho, J. S. Metaplastic breast carcinoma: More than a special type. Nat. Rev. Cancer 14, 147–148 (2014).
Zhu, L. & Chen, K. Clinicopathological features, treatment patterns, and prognosis of squamous cell carcinoma of the breast: An NCDB analysis 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. BMC Cancer 19, 1–9 (2019).
Moreno, A. C. et al. Outcomes after treatment of metaplastic versus other breast cancer subtypes. J. Cancer 11, 1341–1350 (2020).
Lakhani SR El, Schnitt S. J., Tan P. H., van de V. M., WHO Classification of Tumours of the Breast. WHO Press, 2012
Tan, P. H. et al. The 2019 World Health Organization classification of tumours of the breast. Histopathology 77, 181–185 (2020).
Rayson, D., Adjei, A. A., Suman, V. J., Wold, L. E. & Ingle, J. N. Metaplastic breast cancer: Prognosis and response to systemic therapy. Ann. Oncol. 10, 413–419 (1999).
Han, M. L. et al. Metaplastic breast carcinoma: A clinical-pathologic study of 97 cases with subset analysis of response to neoadjuvant chemotherapy. Mod. Pathol. 32, 807–816 (2019).
Bodo, M., Dobrossy, L., Rahoty, P. & Daubner, K. Diagnosis of carcinoma of the breast by aspiration biopsy cytology. Arch. Geschwulstforsch 47, 624–626 (1977).
Hennessy, B. T. et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 69, 4116–4124 (2009).
Taube, J. H. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl Acad. Sci. USA 107, 15449–15454 (2010).
Steinestel, K., Eder, S., Schrader, A. & Steinestel, J. Clinical significance of epithelial-mesenchymal transition. Clin. Transl. Med. 3, 17 (2014).
Ng, C. K. Y. et al. The landscape of somatic genetic alterations in metaplastic breast carcinomas. Clin. Cancer Res. 23, 3859–3870 (2017).
González-Martínez, S. et al. Palacios J Molecular features of metaplastic breast carcinoma: An infrequent subtype of triple negative breast carcinoma. Cancers 12, 1–13 (2020).
Cancer LIA for R on (2019) WHO Classification of Tumors Editorial Board, ed. WHO classification of tumors, 5th edition– Breast tumors.
Martínez-Fernández, P. et al. Comprehensive NGS panel validation for the identification of actionable alterations in adult solid tumors. J. Pers. Med. 11, 1–15 (2021).
Krings, G. & Chen, Y. Y. Genomic profiling of metaplastic breast carcinomas reveals genetic heterogeneity and relationship to ductal carcinoma. Mod. Pathol. 31, 1661–1674 (2018).
Afkhami, M. et al. Mutation and immune profiling of metaplastic breast cancer: Correlation with survival. PLoS One 14, 1–12 (2019).
McCart Reed, A. E. et al. Phenotypic and molecular dissection of metaplastic breast cancer and the prognostic implications. J. Pathol. 247, 214–227 (2019).
Reddy, T. P. et al. A comprehensive overview of metaplastic breast cancer: Clinical features and molecular aberrations. Breast Cancer Res. 22, 1–11 (2020).
Shimoi, T. et al. TERT promoter hotspot mutations in breast cancer. Breast Cancer 25, 292–296 (2018).
Nault, J. C. et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 4, 1–6 (2013).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 5, 405–424 (2015).
Tray, N., Taff, J. & Adams, S. Therapeutic landscape of metaplastic breast cancer. Cancer Treat. Rev. 79, 101878 (2019).
Fallah, Y., Brundage, J., Allegakoen, P. & Shajahan-Haq, A. N. MYC-Driven pathways in breast cancer subtypes. Biomolecules 7, 1–6 (2017).
Reis-filho, J. S. et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J. Pathol. 209, 445–453 (2006).
Gilbert, J. A. et al. Molecular analysis of metaplastic breast carcinoma: high EGFR copy number via aneusomy. Mol. Cancer Ther. 4, 944–951 (2008).
Vendrell, J. A. et al. ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 72, 3593–3606 (2012).
Littlepage, L. E. et al. The transcription factor ZNF217 Is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov. 2, 638–651 (2012).
Bellanger, A. et al. The critical role of the ZNF217 oncogene in promoting breast cancer metastasis to the bone. J. Pathol. 242, 73–89 (2017).
Hayes, M. J., Thomas, D., Emmons, A., Giordano, T. J. & Kleer, C. G. Human cancer biology genetic changes of Wnt pathway genes are common events in metaplastic carcinomas of the breast. Clin. Cancer Res. 14, 4038–4045 (2008).
Acknowledgements
We want to particularly acknowledge the patients and the Biobank HUB-ICO- IDIBELL (PT17/0015/0024) integrated in the Spanish Biobank Network for their collaboration, and also Elvira Purqueras for the management of the tumor samples. Also, we are indebted to the “Biobanc de l’Hospital Infantil Sant Joan de Déu per a la Investigació” Integrated in the Spanish Biobank Network of ISCIII for the sample and data procurement. This manuscript did not receive specific funding.
Author information
Authors and Affiliations
Contributions
P.G., P.C., and I.C. performed genetic tests. A.S., M.C., A.P., J.B.-S., T.S., and S.P. collected and analyzed clinical and anatomo-pathologic information. A.S., P.G., M.C., A.P., J.B.-S., P.C., S.R., A.V., A.F.O., C.F., M.G.G., S.V., R.V.V., T.S., IC, and S.P. integrated and analyzed the clinical data together with molecular data. A.S., P.G., and S.P. wrote the manuscript. All authors reviewed and accepted the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.S. has served as advisor/consultant role for Novartis and Seagen. Payment or honoraria for lectures, presentations, speakers’ bureaus, manuscript writing, or educational events: Eisai, Novartis, Pierre-Fabre. Support for attending meetings and/or travel: Pfizer, Novartis. M.G.G has served as advisor/consultant role for Agendia and Astra-Zeneca. Payment or honoraria for lectures, presentations, speaker’s bureaus, manuscript writing, or educational events: Pfizer, Novartis, Daiichi-Sankyo, and Eisai. Support for attending meetings and/or travel: Roche, Pfizer, Novartis, Lilly, and Daiichi-Sankyo. R.V.V has served as advisor/consultant role for Novartis and Lilly. Payment or honoraria for lectures, presentations, speaker’s bureaus, manuscript writing, or educational events: Novartis, Eisai. Support for attending meetings and/or travel: Novartis, Eisai, and Lilly. S.P. has served as advisor/consultant role for AstraZeneca, Daiichi-Sankyo, Polyphor, Novartis, Seattle Genetics, Roche, Eisai, and Pierre-Fabre.
Ethics approval/consent to participate
Study protocol was approved by the institutional review board at University Hospital of Bellvitge-IDIBELL (Reference: BB20-017). Informed written consent was obtained from all patients alive at the moment of the analysis. All samples were properly coded and anonymized. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Stradella, A., Gargallo, P., Cejuela, M. et al. Genomic characterization and tumor evolution in paired samples of metaplastic breast carcinoma. Mod Pathol 35, 1066–1074 (2022). https://doi.org/10.1038/s41379-022-01017-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-022-01017-7
This article is cited by
-
The mixed subtype has a worse prognosis than other histological subtypes: a retrospective analysis of 217 patients with metaplastic breast cancer
Breast Cancer Research and Treatment (2023)