Abstract
Salivary duct carcinoma (SDC) is an aggressive salivary gland malignancy with poor survival. Approximately 30% SDC harbor HER2 amplification and response to trastuzumab has been reported. However, a systematic approach for HER2 status assessment in this tumor type has not been established. A total of 67 tumor samples were evaluated for HER2 protein overexpression or ERBB2 gene amplification using at least 2 methods: immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), and/or targeted exome next-generation sequencing (NGS). NGS assessed ERBB2 copy number fold change (FC) and total copy number (TCN). HER2 status was first determined by IHC/FISH according to the 2018 ASCO/CAP breast cancer guidelines. FISH results, the “gold standard”, were compared with the NGS results. All (15/15) IHC positive, 35% (6/17) equivocal, and no (0/19) IHC negative SDC were HER2 amplified by FISH. HER2 FISH signal/cell showed a good correlation with FC (Spearman correlation: 0.708, R2: 0.501, p < 0.0001) and TCN (Spearman correlation: 0.763, R2: 0.582, p < 0.0001). Receiver operating characteristics curve estimation showed an area under curve (AUC) of 0.975 for ERBB2 FC. FC cutoff of ≥1.8 corresponded to an accuracy of 95.2% for ERBB2 amplification (Youden’s index: 0.84, sensitivity: 89.47%, specificity: 100%). FC < 1.3 could be reliably classified as ERBB2 not amplified and FC ≥ 1.3 and <1.8 as equivocal. TCN estimation showed AUC of 0.981. TCN cutoff of >6.0 corresponded to an accuracy of 92% for HER2 amplification (Youden’s index: 0.81, sensitivity: 81.2%, specificity: 100%). TCN < 4 could be reliably classified as ERBB2 not amplified and TCN ≥ 4.0 and ≤6.0 as equivocal. FC and TCN were binarized with respective cutoffs of ≥1.8 and ≥6.0 and the proportion of agreement with FISH were 95% and 92%, respectively. The assessment of ERBB2 copy number by NGS is accurate and reliable with FC or TCN nearly equivalent to FISH in identifying HER2 amplified SDC.
Similar content being viewed by others
Introduction
Salivary duct carcinoma (SDC) is a high-grade apocrine adenocarcinoma most frequently found in the parotid gland and represents 2–5% of malignant salivary gland neoplasms1,2. Standard treatment regimen includes surgery, chemoradiation, and androgen receptor (AR) inhibition, but the survival rate remains low and less than 45% patients survive 5 years1. The human epidermal growth factor receptor 2 (HER2) protein, a 185-kDa transmembrane tyrosine-kinase receptor encoded by the HER2 (ERRB2) gene is overexpressed in a variety of malignancies, including approximately 25–30% SDC1,3,4,5,6,7. When activated, the HER2 protein forms heterodimers on the cell surface, which propagates activation of the PI3K and RAS pathways and affects proliferation, survival and angiogenesis8. Therapy with the monoclonal antibody trastuzumab, which causes internalization and downregulation of the HER2 protein, has led to dramatic improvements in disease-free survival in breast cancer patients harboring HER2 amplified tumors9,10,11. Given the pathologic and molecular similarities between SDC and a subset of invasive mammary carcinomas1,7,12,13,14, trastuzumab has been identified as a potential treatment option for HER2 positive SDC. A number of case reports and small case series have demonstrated its effectiveness in the treatment of select SDC patients, especially when combined with other chemotherapeutic drugs15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32. A more recent series of trastuzumab-treated SDC showed most notable responses in cases with HER2 protein overexpression by IHC33. These results support the need for an accurate HER2 scoring system in SDC, which is largely missing at present. The standard methods recommended by the American Society of Clinical Oncology and the College of American Pathologists (ASCO/CAP) for evaluation of HER2 status in breast cancer include detection of the protein overexpression by immunohistochemistry (IHC) and gene amplification by in situ hybridization (ISH)34. The emerging next-generation sequencing (NGS) technologies used in clinical cancer medicine to identify molecular therapeutic targets commonly allow for concurrent detection of copy number alterations (CNA) and can be a cost-effective alternative to multiple single-gene tests. The MSK-IMPACT assay is a clinically validated and U.S. Food and Drug Administration (FDA)-cleared targeted exome sequencing assay that detects point mutations, insertions or deletions, rearrangements, and CNAs35. Using a large set of breast and gastroesophageal carcinomas, MSK-IMPACT was clinically validated to reliably detect ERBB2 amplification and proved to be highly concordant with the traditional methods, IHC and fluorescence in situ hybridization (FISH)36. In this study, we evaluated the HER2 protein expression and ERBB2 gene amplification status in a large cohort of SDC, aiming to assess the concordance between IHC, FISH, and a targeted exome NGS assay in this cancer type.
Methods and materials
Case selection
After obtaining Institutional Review Board approval, the Pathology archives of Memorial Sloan Kettering Cancer Center (MSKCC) were searched for cases with the diagnosis of SDC, which were assessable by at least two of the following assays: HER2 IHC, HER2 FISH, and/or NGS. The diagnostic features of SDC included high-grade histology, apocrine cytology with prominent nucleoli, positive immunolabelling for CK7 and AR, and negative S1007,13,14. A total of 67 SDC were identified, including primary (n = 40) and metastatic (n = 27) formalin-fixed paraffin-embedded (FFPE) tumors diagnosed between 2003 and 2020. The SDC included resection (n = 52), biopsy (n = 14) and cytology (n = 1) samples. Cases were tested for HER2 protein overexpression and/or gene amplification by IHC (n = 63), FISH (n = 55), and NGS (n = 55). HER2 copy number fold change (FC) was evaluated for all cases assessed by NGS and a subset of the NGS profiled cases (n = 44) were also evaluated for the total copy number (TCN). A total of 33 cases were assessed by all four methods. The details of the assays performed are provided in Table 1.
Immunohistochemistry
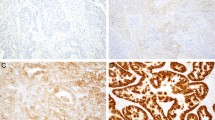
HER2 IHC was performed on 4um FFPE tissue sections using PATHWAY anti-HER-2/neu (4B5) (Ventana, Tucson, AZ) or HercepTest (Dako, Carpinteria, CA). All cases were reviewed by three board-certified anatomic pathologists including one surgical and molecular (DCF), one molecular and breast (DSR), and one molecular and head and neck pathologist (SD). Cases were evaluated using the 2018 ASCO/CAP developed standard system for breast cancer and scored accordingly as depicted in Fig. 134.
Score 0 (negative) shows no staining or incomplete/faint membrane staining in ≤10% of invasive tumor cells. Score 1+ (negative) shows incomplete/faint membrane staining in >10% of invasive tumor cells. Score 2+ (equivocal) shows weak/moderate complete membrane staining in >10% of invasive tumor cells. Score 3+ shows complete, intense membrane staining in >10% of invasive tumor cells. Abbreviations: IHC, immunohistochemistry.
Fluorescence in situ Hybridization
Fluorescence in situ hybridization (FISH) to detect ERBB2 amplification was performed using either the HER2 IQFISH pharmDx (Dako) or PathyVysion HER-2 DNA probe Kit (Vysis, Downers Grove, IL) dual probe kits. A minimum of 50 cancer cells were evaluated in each case by one experienced technologist (TZ) and at least one molecular pathologist (DCF, SD), and the cases were grouped based on the 2018 ASCO/CAP guidelines for breast cancer. (Supplementary Table S1)34.
Copy number assessment by NGS
A hybrid-capture based targeted exome NGS assay (MSK-IMPACT) was performed on 55 cases as previously described, including 29 cases previously published7,35,37. In brief, genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue, and macro-dissection was employed in select cases to enrich for tumor content. The tumor content median was 55% (range 15% to 90%). Either non-neoplastic FFPE tissue or matched peripheral blood were used as source for normal control DNA. Sequencing was performed at a minimum sequencing coverage depth of 200X to interrogate a panel of 341 to 468 cancer-related genes for the presence of point mutations, insertions, deletions and rearrangements. Additionally, the standard clinical pipeline of MSK-IMPACT evaluates more than 1000 intergenic and intronic single-nucleotide polymorphisms to aid in the assessment of copy number alterations. The level of gene copy number gain or loss is calculated from the proportion of sequencing reads at one site relative to another and is referred to as “fold change” (FC). The clinically set threshold for gene amplification is a FC value of 2.0 or greater. Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS) is an allele-specific copy-number analysis pipeline and open-source software for NGS data that is optimized for the MSK-IMPACT assay38. It utilizes heterozygous sites of both a tumor and a normal sample to measure allelic imbalance between the two samples and has the benefit of detecting copy neutral loss of heterozygosity and ploidy. Additionally, this algorithm provides an integer copy number assessment of a specific gene. This value is referred to as for the “total copy number” (TCN).
Statistical analysis for concordance
The optimal cut-offs for FC and TCN relative to the HER2 FISH score which was used as the “gold standard” were determined using receiver operating characteristics (ROC) curve estimation. Spearman correlation was used for evaluation of correlation between non-parametric variables. Kruskal-Wallis test was used for comparison of non-parametric variables between IHC groups. Mann–Whitney test was used to compare NGS FC and TCN based on binarized FISH results. Nam’s score test and McNemar test were used for evaluation of equivalence between assays with an alpha of 0.05.
Results
Comparison of IHC and FISH
Of the 63 cases with IHC results, 21 (33.3%) were positive (score 3+), 22 cases (34.9%) were negative (score 0 or 1+) and 20 cases (31.7%) were equivocal (score 2+; Supplementary Fig. 1). Among 55 cases with both IHC and FISH results, all cases with negative HER2 IHC (scores 0 or 1+, n = 19) were negative for HER2 amplification by FISH. All cases with positive HER2 IHC (3+ score, n = 15) showed HER2 amplification by FISH. Equivocal IHC (2+ score) was detected in 17 cases and 6 (35%) showed HER2 amplification by FISH. Overall, 24 (43.6%) cases were HER2 amplified and 31 (56.4%) were not amplified by FISH. The respective median HER2/CEP17 ratio and median HER2 signals/cell were 5.2 (interquartile range [IQR], 3.6–7.2) and 10.2 (IQR, 6.4–14.7) for amplified, and 1.2 (IQR, 1.1–1.5) and 2.5 (IQR, 2.2–3.0) for not amplified cases. Two cases with a HER2/CEP17 ratio <2 and the average HER2 signals/cell >4.0 and <6.0 were both considered not amplified because they showed negative IHC (scores 0 and 1+, respectively). The HER2/CEP17 ratio and HER2 signal/cell were significantly different between the IHC groups (Kruskal–Wallis P: < 0.0001 for both). The respective median HER2/CEP17 ratio and median HER2 signal/cell were as follows: 1.1 (IQR, 1.0–1.44) and 2.3 (IQR, 2.2–3.0) for IHC negative, 2.8 (IQR, 2.5–4.0) and 6.4 (IQR, 6.2–8.9) for IHC equivocal/FISH amplified cases, 1.3 (IQR, 1.2–1.6) and 2.7 (IQR, 2.5–3.0) for IHC equivocal/FISH not amplified cases, and 5.8 (IQR, 4.2–6.1) and 12.1 (IQR, 10.0–15.6) for IHC positive cases (Fig. 2).
Comparison of NGS and FISH
Out of 42 cases with both FISH and NGS FC results, including 37 with TCN results, 19 (45.2%) cases were amplified, and 23 (54.8%) cases were not amplified by FISH. As determined by the standard clinical pipeline for MSK-IMPACT, the median FC for FISH amplified and FISH not amplified cases were 4.3 (IQR, 2.9–9.2) and −1.0 (IQR, −1.1–1.3; Mann–Whitney p < 0.0001), respectively. HER2 signal/cell, as determined by FISH, showed good correlation with FC (Spearman correlation: 0.708, R2: 0.501, p < 0.0001) (Fig. 3A). Of the 37 cases with TCN data, the respective median TCN for FISH amplified (n = 16) and FISH not amplified cases (n = 21) were 22 (IQR, 10–24) and 1 (IQR, 1–2; Mann–Whitney p < 0.0001). HER2 signal/cell, as determined by FISH, showed good correlation with TCN (Spearman correlation: 0.763, R2: 0.582, p < 0.0001; Fig. 3B). ROC curve estimation analysis (Fig. 3C, Supplementary Table S2) using FISH results as the “gold standard” showed an area under curve (AUC) of 0.975 (95% confidence interval [CI], 0.887–0.995) for ERBB2 copy number FC. A FC cutoff of ≥1.8 corresponded to an accuracy of 95.2% for ERBB2 amplification status (Youden’s index: 0.84, sensitivity: 89.47%, specificity: 100%). At a FC cutoff of ≥1.3 the sensitivity would be 100%, i.e., any case with a FC of <1.3 can be reliably classified as ERBB2 not amplified. FC of ≥1.3 and <1.8 should be considered as equivocal; two FISH amplified cases and five FISH not amplified cases were in this range.
A comparison to FC values shows FC of 1.3–1.7 to be equivocal for amplification (A). A comparison to TCN values shows TCN of 4–6 are equivocal for amplification (B). A ROC curve estimation imputing FISH as the “gold standard” shows both NGS FC and total TCN are reliable predictors of FISH amplification status (C). Abbreviations: FISH, fluorescence in situ hybridization; NGS, next-generation sequencing; FC, fold change; TCN, total copy number; ROC, receiver operating characteristics.
TCN estimation showed AUC of 0.981 (95% CI, 0.907–0.996). The estimated TCN cutoff of >6.0 corresponded to an accuracy of 92% for ERBB2 amplification status (Youden’s index: 0.81, sensitivity: 81.2%, specificity: 100%). With the TCN cutoff of ≥4.0, the sensitivity would be 100%, i.e., any case with a TCN of <4.0 can be reliably classified as ERBB2 not amplified. TCN of ≥4.0 and ≤6.0 should be considered equivocal; three FISH amplified and three FISH not amplified cases were in this range.
TCN and FC showed an excellent correlation (Spearman correlation: 0.94, R2: 0.712, P: <0.0001). TCN and FC performed similarly in the assessment of ERBB2 amplification status; one FISH not amplified case was negative by TCN 2 but equivocal by FC 1.4; one FISH amplified case was positive by FC 1.8 and equivocal by TCN 4; all other cases were concordant. To compare the performance of NGS with FISH, the FC and TCN were binarized with cutoffs of ≥1.8 and >6.0, respectively. After binarization, an equivalence comparison of NGS to FISH was made; the proportion of agreement for FC and FISH was 0.95 (Nam score: −0.05, McNemar P: 0.15) and the proportion of agreement for TCN and FISH was 0.92 (Nam score: −0.08, McNemar P: 0.08), demonstrating that the accuracy of NGS assessment of ERBB2 copy number status using either FC or TCN is nearly equivalent to FISH.
Comparison of NGS and IHC
Among 50 cases with both IHC and NGS results available, 18 cases (36%) were IHC negative (score 0 or 1+), 15 cases (30%) were IHC equivocal (score 2+) and 17 (34%) cases were IHC positive (score 3+). Out of 17 IHC positive cases, 16 (94.1%) were positive and one (5.9%) was equivocal by FC (NGS). Out of 15 cases with IHC equivocal cases, 4 (26.7%) were amplified, 6 (40%) were not amplified, and 5 (33.3%) were equivocal by FC. Out of 5 FC equivocal cases with available FISH results, 1 case was ERBB2 amplified. FC and TCN were significantly different between the IHC groups (Kruskal–Wallis, p < 0.0001 for both). The respective median FC and median TCN were −1.1 (IQR, −1.1 – −1.0) and 1 (IQR, 1–1) for negative IHC, 2.9 (IQR, 1.8–3.1) and 8 (IQR, 5–14) for equivocal IHC/FISH amplified cases, 1.4 (IQR, 0.0–1.5) and 2 (IQR, 1–3) for equivocal IHC/FISH not amplified cases, and 4.7 (IQR, 4.1–8.1) and 24 (IQR, 19–29) for positive IHC (Fig. 4).
Comparison of IHC, FC, and TCN and the proposed HER2 testing algorithm in SDC
Based on 33 cases tested by all four methods, the proportion of equivocal cases defined by IHC, FC, and TCN were 10 (30.3%), 5 (15.2%), and 5 (15.2%), respectively. FC and TCN performed similarly in predicting the HER2 status and either method was superior to IHC alone (Fig. 5). A summary of HER2 testing recommendations is depicted in Fig. 6. In brief, a second method would be needed for the equivocal categories including IHC 2+, FC ≥ 1.3 or <1.8, or TCN ≥ 4 or <7.
The order of reflex testing is interchangeable and can be based on the availability of HER2 testing assay. *Further studies are needed for validation. Abbreviations: SDC, salivary duct carcinoma; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; NGS, next-generation sequencing; FC, fold change; TCN, total copy number; CN, copy number.
Discussion
HER2 positivity is a well-established biomarker for a variety of cancer types due to the vastly improved overall survival rates with anti-HER2 targeted therapy. A significant subset of SDC cases are HER2 positive and preliminary data have demonstrated the efficacy of anti-HER2 targeted therapy, such as trastuzumab, in these patients15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32. Assembling a relatively large cohort of SDC, we evaluated four testing methods to identify HER2 positive cases, analyzed their concordance, and utilized those results to compile a consistent HER2 scoring system for SDC effective for IHC, FISH, and NGS.
The IHC and FISH results were interpreted according to the 2018 ASCO/CAP guidelines for HER2 testing in breast cancer and this conventional testing approach might be sufficient in most clinical settings. However, the evolvement of genomic medicine and the increasing clinical need for comprehensive molecular testing mandate the development of tissue-efficient alternatives to single-gene testing methods. NGS has proven to be useful in detecting the ERBB2 amplification in other cancer types including breast, endometrial, gastroesophageal, and colorectal carcinomas36,39,40, and this approach could also be applied in SDC.
Based on the ROC curve estimation analysis of our SDC data, we found that the optimal threshold for ERBB2 amplification in SDC would be FC ≥ 1.8, which is lower than the standard cut off for gene amplification by MSK-IMPACT assay, FC ≥ 235. In addition, among the cases in the equivocal range, FC 1.3–1.7, we identified two HER2 positive cases. This observation is consistent with the prior study on ERBB2 amplification in breast cancer published by our group, where the optimal sensitivity (95%) and specificity (100%) could also be achieved below FC 2 i.e., an amplified case could show FC ≥ 1.5 but <2. These were typically cases with relatively low tumor content in the tested sample or those showing intratumoral heterogeneity in HER2 expression36.
Although 15.2% of cases would fall into the equivocal FC range and require alternate testing (i.e., FISH), a proportion of these cases is lower than equivocal cases tested by HER2 IHC supporting the superiority of NGS to IHC in identifying the truly HER2 positive and negative cases. Next, we utilized TCN data generated by FACETS to assess ERBB2 amplification. Despite FACETS’ current lack of widespread clinical use and undefined amplification thresholds, its inclusion in this study represented an opportunity to expand the methods available for identifying HER2 positive cases. In a prior study using MSK-IMPACT for evaluation of ERBB2 amplification status (prior to use of FACETS) TCN 6 was suggested as the cut off for HER2 amplification35. However, in our data set this cut off was associated with a slightly lower accuracy rate (89%) and we found that TCN 7 can reliably detect HER2 positive and TCN < 4 identifies HER2 negative cases.
The scoring schemes utilizing IHC, FISH, and NGS were in complete concordance when analyzed. Although there were equivocal cases for IHC and NGS that were only definitive using FISH, there were no contradictions in returned results and most cases could be definitively scored using any of the three systems/four methods. The order of testing is interchangeable and equivocal results from any assay would require a further testing by an alternate method. In terms of NGS evaluation of ERBB2 amplification status, the performance of FC and TCN is similar and either method can be used for this purpose.
This study has several strengths. First, given the rarity of SDC in the general population, the large cohort of samples included in this study provides a greater insight into the reliability of testing methods. Second, by comparing the two most commonly applied methods for determining HER2 status in SDC with two emerging NGS methods and finding them concordant, this study demonstrates that NGS can be an effective stand-alone method for this purpose. This is beneficial because NGS, in addition to providing the ERBB2 amplifications status, also provides information on other therapeutic and prognostic biomarkers. Third, our study examines an investigational bioinformatics algorithm (FACETS) method for determining the ERBB2 gene amplification and suggests amplification thresholds for future clinical use. This study also has several limitations. A limited number of cases were tested by all four testing modalities and larger number of cases and/or additional datasets would be needed for further validation of the suggested cut-off values. Furthermore, the outcome data relative to the ERBB2 status for the cases utilized in this study are not available. In order to determine the effectiveness of the suggested scoring systems, the outcome of anti-HER2 treatments must be compared to the ranges identified as HER2 positive. Although this study does provide the framework to apply the existing methods in a consistent manner, it does not make a determination that the ranges within each scoring system are accurate predictors of drug response.
In summary, we examined the utility of four methods to detect HER2 positive SDC cases. We found that in addition to conventional IHC and FISH, NGS can be used as a single method for ERBB2 amplification detection in the majority of cases. While these data may help identify SDC patients more likely to respond to anti-HER2 therapy, clinical studies focused on the response to targeted therapy in SDC patients are needed for the ultimate clinical validation of the proposed HER2 testing scoring schemes.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Chiosea, S. I. et al. Molecular characterization of apocrine salivary duct carcinoma. Am. J. Surg. Pathol. 39, 744–752 (2015).
Guzzo, M. et al. Major and minor salivary gland tumors. Crit. Rev. Oncol. Hematol. 74, 134–148 (2010).
Williams, M. D. et al. Genetic and expression analysis of HER-2 and EGFR genes in salivary duct carcinoma: empirical and therapeutic significance. Clin. Cancer Res. 16, 2266–2274 (2010).
Sugano, S. et al. Immunohistochemical study of c‐erbB‐2 oncoprotein overexpression in human major salivary gland carcinoma: an indicator of aggressiveness. Laryngoscope 102, 923–927 (1992).
Skalova, A., Starek, I., Kučerová, V., Szépe, P. & Plank, L. Salivary duct carcinoma-a highly aggressive salivary gland tumor with HER-2/neu oncoprotein overexpression. Pathol-Res. Pract. 197, 621–626 (2001).
Press, M. F. et al. Amplification and overexpression of HER-2/neu in carcinomas of the salivary gland: correlation with poor prognosis. Cancer Res. 54, 5675–5682 (1994).
Dogan, S. et al. The repertoire of genetic alterations in salivary duct carcinoma including a novel HNRNPH3-ALK rearrangement. Hum. Pathol. 88, 66–77 (2019).
Kramer-Marek, G. & Oyen, W. J. Targeting the human epidermal growth factor receptors with immuno-PET: imaging biomarkers from bench to bedside. J. Nucl. Med. 57, 996–1001 (2016).
Di Leo, A. et al. Phase III, double-blind, randomized study comparing lapatinib plus paclitaxel with placebo plus paclitaxel as first-line treatment for metastatic breast cancer. J. Clin. Oncol. 26, 5544–5552 (2008).
Konecny, G. E. et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 66, 1630–1639 (2006).
Seidman, A. D. et al. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: final results of Cancer and Leukemia Group B protocol 9840. J. Clin. Oncol. 26, 1642–1649 (2008).
Dalin, M. G. et al. Comprehensive molecular characterization of salivary duct carcinoma reveals actionable targets and similarity to apocrine breast cancer. Clin. Cancer Res. 22, 4623–4633 (2016).
Di Palma, S. et al. Salivary duct carcinomas can be classified into luminal androgen receptor‐positive, HER2 and basal‐like phenotypes. Histopathology 61, 629–643 (2012).
Williams, L. et al. Salivary duct carcinoma: the predominance of apocrine morphology, prevalence of histologic variants, and androgen receptor expression. Am. J. Surg. Pathol. 39, 705–713 (2015).
Bergamini C., Cavalieri S., Sanguineti G., Farneti A., Licitra L. Treatment of HER2+ metastatic salivary ductal carcinoma in a pregnant woman: a case report. Oxf Med Case Reports 2019, omz102. eCollection (2019)
van Boxtel, W. et al. Combination of docetaxel, trastuzumab and pertuzumab or treatment with trastuzumab-emtansine for metastatic salivary duct carcinoma. Oral Oncol. 72, 198–200 (2017).
De Block, K. et al. Metastatic HER-2-positive salivary gland carcinoma treated with trastuzumab and a taxane: a series of six patients. Acta Clin. Belg. 71, 383–388 (2016).
Falchook, G. S., Lippman, S. M., Bastida, C. C. & Kurzrock, R. Human epidermal receptor 2–amplified salivary duct carcinoma: Regression with dual human epidermal receptor 2 inhibition and anti–vascular endothelial growth factor combination treatment. Head Neck 36, E25–E27 (2014).
Gibo, T. et al. Targeted therapy with trastuzumab for epidermal growth factor receptor 2 (HER2)‑positive advanced salivary duct carcinoma: a case report. Mol. Clin. Oncol. 11, 111–115 (2019).
Iqbal, M. S., Shaikh, G., Chatterjee, S., Cocks, H. & Kovarik, J. Maintenance therapy with trastuzumab in her2 positive metastatic parotid ductal adenocarcinoma. Case Rep. Oncol. Med. 2014, 162534 (2014).
Kadowaki, S. et al. Complete response to trastuzumab-based chemotherapy in a patient with human epidermal growth factor receptor-2-positive metastatic salivary duct carcinoma ex pleomorphic adenoma. Case Rep. Oncol. 6, 450–455 (2013).
Kaidar-Person, O., Billan, S. & Kuten, A. Targeted therapy with trastuzumab for advanced salivary ductal carcinoma: case report and literature review. Med. Oncol. 29, 704–706 (2012).
Krishnamurthy, J. et al. Salivary duct carcinoma responding to trastuzumab‐based therapy: case report and review of the literature. Head Neck 35, E372–E375 (2013).
Limaye, S. A. et al. Trastuzumab for the treatment of salivary duct carcinoma. Oncologist 18, 294–300 (2013).
Locati, L. et al. Herceptin plus chemotherapy in relapsed and/or metastatic salivary gland cancer. Oral Oncol. 41, 97–98 (2005).
Nabili, V., Tan, J. W., Bhuta, S., Sercarz, J. A. & Head, C. S. Salivary duct carcinoma: a clinical and histologic review with implications for trastuzumab therapy. Head Neck 29, 907–912 (2007).
Nashed, M. & Casasola, R. J. Biological therapy of salivary duct carcinoma. J. Laryngol. Otol. 123, 250–252 (2009).
Park, J. C. et al. Exceptional responses to pertuzumab, trastuzumab, and docetaxel in human epidermal growth factor receptor‐2 high expressing salivary duct carcinomas. Head Neck 40, E100–E106 (2018).
Perissinotti, A. J. et al. The role of trastuzumab in the management of salivary ductal carcinomas. Anticancer Res. 33, 2587–2591 (2013).
Prat, A. et al. Successful treatment of pulmonary metastatic salivary ductal carcinoma with trastuzumab-based therapy. Head Neck 30, 680–683 (2008).
Swed B. L., Cohen R. B., Aggarwal C. Targeting HER2/neu oncogene overexpression with ado-trastuzumab emtansine in the treatment of metastatic salivary gland neoplasms: a single-institution experience. JCO Precision Oncol. 3, https://doi.org/10.1200/PO.18.00351 (2019)
Ueki, Y. et al. Pathological response of salivary duct carcinoma to trastuzumab and docetaxel therapy. Int. Cancer Conf. J. 5, 150–153 (2016).
Hanna, G. J. et al. The benefits of adjuvant Trastuzumab for HER‐2‐positive salivary gland cancers. Oncologist 25, 598–608 (2020).
Wolff, A. C. et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. Arch. Pathol. Lab. Med. 142, 1364–1382 (2018).
Cheng, D. T. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Ross, D. S. et al. Next-generation assessment of human epidermal growth factor receptor 2 (ERBB2) amplification status: clinical validation in the context of a hybrid capture-based, comprehensive solid tumor genomic profiling assay. J. Mol. Diagn. 19, 244–254 (2017).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Robinson, C. L. et al. Detection of ERBB2 amplification in uterine serous carcinoma by next-generation sequencing: an approach highly concordant with standard assays. Mod. Pathol. 34, 603–612 (2021).
Cenaj, O., Ligon, A. H., Hornick, J. L. & Sholl, L. M. Detection of ERBB2 amplification by next-generation sequencing predicts HER2 expression in colorectal carcinoma. Am. J. Clin. Pathol. 152, 97–108 (2019).
Acknowledgements
We gratefully acknowledge the members of the MSKCC Molecular Diagnostics Service of the Department of Pathology. This work has been supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748.
Author information
Authors and Affiliations
Contributions
D.C.F., A.M.B., and S.D. performed study concept and design; D.C.F., A.M.B., A.L.H., and S.D. were responsible for acquisition of data; D.C.F., A.M.B., A.S.M., T.Z., D.S.R., and S.D. interpreted and analyzed the data; A.M.B. performed statistical analyses; T.Z. provided technical support; D.C.F., A.M.B., and S.D. wrote the manuscript; D.C.F., A.M.B., A.S.M., A.L.H., M.E.A., D.S.R., and S.D. performed review and revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
No competing financial interests exist for all contributory authors. Research reported in this publication was supported by the Memorial Sloan Kettering Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ferguson, D.C., Momeni Boroujeni, A., Zheng, T. et al. ERBB2 amplification status in 67 salivary duct carcinomas assessed by immunohistochemistry, fluorescence in situ hybridization, and targeted exome sequencing. Mod Pathol 35, 895–902 (2022). https://doi.org/10.1038/s41379-021-00999-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00999-0
This article is cited by
-
Oncocytoid Salivary Tumors: Differential Diagnosis and Utility of Newly Described Immunohistochemistry
Head and Neck Pathology (2024)