Abstract
Classification of cancers by tissue-of-origin is fundamental to diagnostic pathology. While the combination of clinical data, tissue histology, and immunohistochemistry is usually sufficient, there remains a small but not insignificant proportion of difficult-to-classify cases. These challenging cases provide justification for ancillary molecular testing, including high-throughput DNA methylation array profiling, which promises cell-of-origin information and compatibility with formalin-fixed specimens. While diagnostically powerful, methylation profiling platforms are costly and technically challenging to implement, particularly for less well-resourced laboratories. To address this, we simulated the performance of “minimalist” methylation-based tests for cancer classification using publicly-available and internal institutional profiling data. These analyses showed that small and focused sets of the most informative CpG biomarkers from the arrays are sufficient for accurate diagnoses. As an illustrative example, one classifier, using information from just 53 out of about 450,000 available CpG probes, achieved an accuracy of 94.5% on 2575 fresh primary validation cases across 28 cancer types from The Cancer Genome Atlas Network. By training minimalist classifiers on formalin-fixed primary and metastatic cases, generally high accuracies were also achieved on additional datasets. These results support the potential of minimalist methylation testing, possibly via quantitative PCR and targeted next-generation sequencing platforms, in cancer classification.
Similar content being viewed by others
Introduction
Tissue-of-origin information is important for cancer diagnosis. While the current paradigm that combines clinical data, histology, and immunohistochemistry (IHC) permits the classification of the vast majority of cancers, difficult cases remain. These include cancers of unknown primaries (CUPs) [1] and other challenging differentials from pathology subspecialties that provide impetus for ancillary molecular testing, including DNA methylation array profiling. As examples of diagnostic profiling, Moran et al. [2] developed a pan-cancer classifier called EPICUP, which provided a diagnosis for 87% of 216 CUPs; interestingly, among patients with EPICUP diagnoses, those who received tumor-specific therapies survived longer than those who received empiric therapies for CUPs; Capper et al. [3] published a methylation array-based classifier for CNS tumors (molecularneuropathology.org); strikingly, predictions from that classifier changed 12% of the original neuropathology diagnoses in an independent validation cohort.
While biologically informative and diagnostically useful [2,3,4,5,6,7], tests based on methylation array profiling are costly and technically challenging to implement as a new platform. In particular, investment in new equipment (e.g., Illumina instruments), reagents, and additional capacity for testing may be required, which essentially restricts such endeavors to major centers or reference laboratories. For many smaller clinical laboratories, where limited resources are stretched thin by existing and anticipated mandates, investments in methylation array profiling would be unrealistic.
In this paper, we propose that cancer tissue-of-origin testing by DNA methylation can be greatly simplified. Our hypothesis is that high accuracies can be achieved using small and focused sets of the most informative CpG markers (rather than all probes) on an array, and that as a consequence of this, testing can be dramatically scaled down and adapted for lower throughput, potentially less expensive, and more readily-available platforms—such as targeted next-generation sequencing (NGS) and quantitative PCR. To evaluate this proposition, we first identified the most informative diagnostic CpG probes by analyzing methylation profiling data from The Cancer Genome Atlas (TCGA) Network, and then assessed the diagnostic performances of “minimalist” classifiers, based on small numbers of these probes, on both fresh primary and formalin-fixed primary and metastatic cancer samples from multiple sources, predominantly cases with known primary origins. To explore the applicability of the minimalistic classifier methodology to a sequencing-based analysis platform, a classifier for high-throughput methylation sequencing (methyl-seq) data was custom-built and applied to a 15-sample dataset. The results, implications, and limitations are presented below.
Materials and methods
Gathering of publicly-available DNA methylation array data
TCGA Human Methylation 450K array data from fresh primary cases were downloaded from the Broad Institute GDAC website (https://gdac.broadinstitute.org). Since esophageal adenocarcinomas (EADs) are distinct from esophageal squamous cell carcinomas (ESCCs), we separated the esophageal carcinoma (ESCA) dataset into ESCCs and EADs. Since EADs closely resemble the chromosome instability subtype of gastric adenocarcinomas (STADs) at the molecular level [8,9,10], we combined EADs and STADs into a single gastric/esophageal adenocarcinoma (GEAD) class. Since only a few ovarian serous carcinomas (OV) were profiled using the 450K array, OV was not included in this study. The TCGA fresh primary dataset is detailed in Supplementary Table 1 (n = 8537, 28 cancer classes).
Methylation array data from formalin-fixed paraffin embedded (FFPE) primary and metastatic samples were obtained from TCGA (via the Broad GDAC website), the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) [3, 11,12,13,14,15,16,17,18,19,20,21], and institutional collaborators (see next section). The GEO dataset included select CNS tumors profiled by Capper et al. [3]: glioblastomas [22], low-grade gliomas corresponding to types studied by TCGA [23], metastatic melanomas, and primary CNS diffuse large B-cell lymphomas. The various datasets along with institutional lung cancer primary and metastasis cases (see next section) are recombined into the FFPE primary (Supplementary Table 2; n = 1016, 12 cancer types), FFPE brain metastasis (Supplementary Table 3; n = 142, 4 cancer types including three CUPs), and FFPE lymph node metastasis cohorts (Supplementary Table 4; n = 29, 2 cancer types) for classifier training and validation.
Additional publicly-available methylation array data (450K and 27K) for the gastric versus pancreatic adenocarcinoma differential were obtained from GEO [24,25,26,27,28,29,30] and the International Cancer Genome (ICGC; https://dcc.icgc.org/) for training and validation. Please see Supplementary Table 5 (n = 854 fresh frozen and archival cases, two cancer types).
For this study, the gold-standard reference for tumor type/primary origin is that specified by TCGA, ICGC, study authors, or UHN pathologists/research collaborators (for internal data).
Since the CpG sites included in different datasets varied, the lists of available probes were “downsized” in some analyses, such that only highly-ranked probes (see below) shared across all included datasets for each analysis were used for classifier training and testing. Beta values from methylation datasets were converted to M values [31] prior to feature ranking, and random forest (RF) model training and validation. Probes containing exclusively NAs or infinite M values and poorly performing probes identified in a recent published study [32] were removed.
Institutional DNA methylation array and methyl-seq data
Archival tissues from 133 FFPE primary lung adenocarcinomas, 39 lung squamous cell carcinomas, and 14 lung adenocarcinoma brain metastases were obtained from the Department of Pathology at UHN. We also obtained FFPE tissues from a cohort of 15 patients enrolled in the REACT study that included one bladder cancer, two cervical and endocervical cancers, six colorectal cancers, two lung adenocarcinomas, two melanomas, and two uterine corpus endometrial carcinomas. DNA methylation array analyses were performed at the Ontario Institute of Cancer Research as follows: 250 ng of DNA were treated using the Illumina FFPE DNA Restore Kit, hybridized to the Infinium EPIC methylation array, and scanned using an iScan instrument; raw data were processed using the Bioconductor package minfi v1.30 for normalization (Illumina method) and extraction of methylation values.
Methyl-seq analyses were also performed in parallel in the 15-patient cohort (REACT study) using a second aliquot of 500 ng of DNA from each sample; after bisulfite conversion, the DNA was subjected to library construction and targeted capture using the Illumina TruSeq Methyl Capture EPIC Library Prep Kit (targeting ~800,000 CpG sites), and followed by sequencing (Illumina NextSeq550, 70 million × 100 paired-end reads); methylation intensities were calculated with the Bismark v0.20.0 pipeline.
Identifying the most informative CpG features using TCGA cases
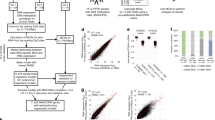
Seventy percent of randomly selected cases from TCGA dataset (training dataset, n = 5962 involving 28 cancer types, Supplementary Table 1) were used to identify the most informative diagnostic probes via two types of feature ranking studies. The first type (see Fig. 1a, henceforth X vs. all) selected for markers best able to distinguish one cancer type from all other cancer types. The second approach (see Fig. 1b, henceforth pairwise differential) selected for markers best able to distinguish one cancer type from a second cancer type, for all possible pairwise combinations.
a To illustrate the feature ranking methods, consider a hypothetical differential consisting of five cancer types (T1 through T5). The X vs. all approach seeks to identify the markers that distinguish each tumor type from all other tumor types. In this illustration, the corresponding analyses result in five feature rank lists that prioritize probes for training X vs. all classifiers. The pairwise differential approach aims to identify the best markers for differentiating each possible tumor pair (e.g., T1 vs. T2). These analyses result in ten prioritized lists, which are used in the training of pairwise classifiers. One disadvantage of the pairwise approach is that the numbers of analyses increase dramatically with numbers of cancer classes (i.e., n = # cancer classes, # analyses = n(n − 1)/2). Hybrid classifiers start with X vs. all lists, and depending on classifier performance, specific pairwise lists corresponding to difficult differentials (red; in this example, T3 vs. T4) are subsequently added—ideally, striking a balance between minimalism and accuracy. The ranked lists can also be combined in various other combinations, depending on the specific differential being considered; Table 1 shows examples of various minimalist classifiers in this study, all developed based on highly-ranked probes from TCGA analyses. b Example of a highly informative CpG marker (cg24727122, OSM, chr22: 30662972) from the X vs. all analysis that accurately separates acute myeloid leukemias (n = 135, red circle) from the other 27 TCGA cancer types (n = 5827; AUC = 1.00). c Heat map for the AUCs of the top-ranked CpG biomarker identified in the 378 pairwise differential analyses; the top AUCs were above 0.98 for most pairwise differentials in TCGA training cases (see Supplementary Table 8 for the numerical values). ACC adrenocortical carcinoma, BLCA bladder carcinoma, BRCA breast invasive carcinoma, CESC cervical and endocervical cancers, CHOL cholangiocarcinoma, CORE colorectal adenocarcinoma, DLBC diffuse large B-cell lymphoma, ESCC esophageal squamous cell carcinoma, GBMLGG glioma (glioblastoma and low-grade glioma), GEAD gastric and esophageal carcinoma, HNSC head and neck squamous cell carcinoma, KIPAN pan-kidney cohort (clear cell, chromophobe, and papillary renal cell carcinoma), LAML acute myeloid leukemia, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, MESO mesothelioma, PAAD pancreatic adenocarcinoma, PCPG pheochromocytoma and paraganglioma, PRAD prostate adenocarcinoma, SARC sarcoma, SKCM skin cutaneous melanoma, TGCT testicular germ cell tumor, THCA thyroid carcinoma, THYM thymoma, UCEC uterine corpus endometrial carcinoma, UCS uterine carcinosarcoma, UVM uveal melanoma.
For 28 cancer types, there were 28 possible analyses for the X vs. all, and 378 possible analyses for pairwise differentials. For each analysis, all probes on the array were ranked in descending order by areas under the receiver operator curves (ROC AUCs). Probes that have the same AUC were further ranked from most significant to least by unadjusted p values from Wilcoxon tests. For each analysis, we also calculated the difference between the median beta values between the analyzed groups (e.g., cancers X and Y) for each probe; probes with differences of <0.1 by median beta were removed after the ROC/Wilcoxon analyses. The number of remaining probes with AUCs ≥0.9 and the AUC of the highest ranked probe from each analysis were recorded.
Evaluation of pan-cancer random forest (RF) classifiers using TCGA fresh primary cases
For fresh primary cancers from TCGA, RF classifiers were trained using TCGA training cases (n = 5962, 28 cancer types). Series of sequentially larger models were trained based on information from the top-one, top-two, top-three, etc., ranked probes from the (1) 28 X vs. all, (2) 378 pairwise differential, or (3) a combination (or hybrid) of the two types of feature ranking analyses (Fig. 1a). The hybrid models was developed manually; we started with the 28 X vs. all lists, and additional pairwise lists were later incorporated corresponding to specific difficult differentials, defined as those with more than five total errors or an error rate of >0.05 from the validation confusion matrix for the smallest X vs. all model.
The remaining 30% of TCGA cases were used for classifier testing (validation dataset, n = 2575, 28 cancer types, Supplementary Table 1). We recorded the overall accuracies and numbers of unique features used for sequentially larger models, to evaluate the trade-off between the reduction in the number of features used and the increase in classification errors. In addition, the sensitivity, specificity, positive and negative predictive values (PPVs and NPVs), and balanced accuracies for each of the 28 tumor classes for specific models were recorded. t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction was used to visualize cancer clusters for the smallest hybrid model.
Since RF is an ensemble machine learning approach, where classifiers consist of collections of decision trees (n = 500 in this study), we recorded the proportion of trees that rendered each consensus call (i.e., the most common prediction from the 500 trees) as a measure of prediction confidence, for the smallest hybrid model.
Using published methylation-based tumor purity estimates for TCGA cases [33] and performance data for the smallest hybrid pan-cancer TCGA classifier, we assessed the association between (1) tumor purity and diagnostic accuracy using the Wilcoxon test, and (2) purity and confidence levels using the Spearman test in TCGA validation cases.
Evaluation of minimalist classifiers on FFPE primary and FFPE metastasis samples
Models for classifying FFPE primary and metastatic cancers are summarized in Table 1 (along with the smallest hybrid pan-cancer classifier). Highly-ranked probes used for classification were those previously identified from TCGA pairwise differential analyses (Fig. 1). Briefly, two models were trained to classify FFPE primary cases. One model was trained using TCGA training cases only, and another was trained using a combination of TCGA training and 70% of randomly selected FFPE primary cases. Both were validated using the remaining 30% of FFPE primary cases (see Table 1). A single model was trained using 70% of randomly selected FFPE brain metastasis cases and validated on the remaining 30% of FFPE brain metastasis cases (Table 1). t-SNE dimensionality reduction was used to visualize brain metastasis cancer clusters for the brain metastasis model.
For FFPE lymph node metastases, we determined the optimal cutoff for a single highly-ranked probe from the TCGA breast carcinoma versus melanoma differential via the Youden method, and validated this cutoff on a separate set of TCGA primary and FFPE lymph node metastasis cases (Table 1).
Evaluation of minimalist classifiers on gastric and pancreatic cancers
For gastric versus pancreatic adenocarcinomas, we determined the optimal cutoff for a single highly-ranked probe from the TCGA gastric/esophageal versus pancreatic adenocarcinoma differential via the Youden method, and validated this cutoff using independent cases from (1) TCGA and (2) ICGC and GEO (Table 1).
Development of a minimalistic classifier for methyl-seq data
CpG sites in the methyl-seq dataset were annotated based on EPIC array annotations; CpG sites with >10% missing values and sites not present on the 450K array were removed, resulting in a filtered methyl-seq dataset containing 123,783 methylation sites. A classifier was trained using 24 selected CpG sites and methylation values from TCGA fresh primary and FFPE primary methylation array data (Table 1). This classifier was tested on 15 cases profiled in parallel on methyl-seq and DNA methylation array, and the correlations of the methylation values for the 24 CpG sites between the two platforms for the 15 cases were measured.
Gene enrichment analyses
A list of unique genes was generated from the top-10 ranked CpG sites from 378 pairwise differential lists from TCGA feature ranking analyses. The genes were analyzed for enrichment using the Enrichr tool [34, 35], including for Gene Ontology (GO) biological processes, cellular components, and molecular functions.
Results
There are large numbers of highly informative CpG sites for cancer classification
TCGA training dataset (70% of randomly selected cases, n = 5962, 28 cancer types, Supplementary Table 1) was used to identify the most informative diagnostic probes, which form the basis for all classifiers (including those for FFPE primary and metastasis samples, and samples tested via methyl-seq) in this study. The feature ranking strategies are described in “Methods” and Fig. 1. For 28 cancer classes, the analyses produced 28 rank lists of probes that distinguish each cancer class from the other 27 classes (X vs. all), and 378 rank lists of probes that distinguish one cancer class from another for all possible pairwise differentials. For each differential, probes on the array were ranked by AUCs in ROC analyses and p values.
Several trends were apparent. First, most analyses identified large numbers of highly informative CpG sites for their respective differentials, defined as those with AUCs ≥0.90 (Fig. 1b, c and Supplementary Tables 6–8); however, there were a few exceptions; in particular, there were relatively few highly informative probes able to separate some types of squamous cell carcinomas (SCCs). Second, the pairwise differential rankings studies generally identified larger numbers of features with better AUCs than the X vs. all (Supplementary Tables 6–8); this is not surprising since biomarkers that work well for separating cancer T1 from T2 may fail to separate T1 from cancer types other than T2. Finally, many top-ranked probes from the analyses had AUCs at or approaching 1.00 across training cases (Fig. 1b, c and Supplementary Tables 6, 8), implying that small numbers of these individual probes could be sufficient for accurate diagnosis.
Small numbers of highly informative CpG sites are sufficient for accurate classification of TCGA cases
Based on this, RF machine learning models, using information limited to small numbers of the most informative probes from feature ranking analyses, were developed using TCGA training cases (Figs. 1, 2 and “Methods”). Trained classifiers were tested on an independent TCGA validation cohort (n = 2575, 28 cancer classes, Supplementary Table 1).
Each point represents a single classifier from the (a) X vs. all, (b) pairwise differential, or (c) hybrid series. The smallest models are indicated; the smallest hybrid model is shown in greater detail in Fig. 3. The colored trend lines for the plots are rectangular hyperbolas with asymptotes (maximum predicted accuracies) of 94.9% (blue), 96.8% (red), and 96.9% (green) for the X vs. all, pairwise, and hybrid series, respectively. Overall, little additional accuracy is gained by the incorporation of additional highly-ranked probes.
We began by studying the correlation between the numbers of features included in RF models and classification accuracies. For this, we trained sequentially more complex classifiers using the top-one, top-two, top-three, etc. features from the (1) X vs. all, (2) pairwise differential, and (3) hybrid feature rank lists. The accuracies of individual models were plotted against the number of features included (Fig. 2). The smallest models used only a single top-ranked CpG marker from each feature rank list. As examples, the smallest X vs. all classifier included just 28 unique probes (one for each cancer class, listed in Supplementary Table 9), and achieved an overall accuracy of 88.5% on independent validation; the smallest pairwise differential model used information from 283 unique probes (less than the 378 pairwise differential lists because some of the top-ranked features were duplicated across two or more differentials, Supplementary Table 10), and correctly predicted 96.4% of diagnoses; the smallest hybrid model included 53 unique probes (see “Methods”, and Supplementary Table 11), and had an accuracy of 94.5%. Figure 2 shows that with the incorporation of additional highly-ranked features, there were only slight improvements in classifier accuracies for the X vs. all and hybrid series, and no definite improvements in accuracies for the pairwise series. Critically, this suggests that minimalist classifiers based on small numbers of the best features are (almost) as accurate as more complex models that use information from the entire array, at least in the fresh primary pan-cancer setting.
Figure 3 highlights the smallest hybrid model in greater detail, as a simulated example of a pan-cancer test (e.g., conceivably, implemented as a targeted next-generation sequencing test) that balances accuracy against simplicity. The t-SNE plot (Fig. 3a), based on the 53 probes from that model, showed generally well-separated cancer clusters, with the exception of some SCC classes. Concordantly, the heat map of the confusion matrix (Fig. 3b and Supplementary Table 12) and performance metrics (Tables 1, 2) demonstrated accurate discrimination of most histologic types, with the exception of SCCs from different anatomic sites.
a t-distributed stochastic neighbor embedding (t-SNE) plot for the smallest hybrid model based on information from 53 unique CpG sites shows excellent separation of cancer classes. b Heat map for the confusion matrix for the smallest hybrid model; see Supplementary Table 12 for the numbers of cases in each cell. c The relationship between classifier confidence and accuracy: the numbers of cases/percentages of validation set/accuracies for the high, moderate, and low confidence groups are 994 cases/39% of validation cases/100% accuracy, 1147/45% of cases/98% accuracy, and 434/17% of cases/73% accuracy, respectively. d Correctly classified cases have statistically higher tumor purities compared with incorrectly classified cases (Wilcoxon test p value = 2.8 × 10−4), although the difference in the distributions is modest. e Density scatter plot showing a direct correlation between purity and prediction confidence (Spearman rho = 0.19); many TCGA cases have fairly high purities (>50%) and many have high confidence predictions. Conceivably, these 53 probes could be quantitatively evaluated via next-generation sequencing. ACC adrenocortical carcinoma, BLCA bladder carcinoma, BRCA breast invasive carcinoma, CESC cervical and endocervical cancers, CHOL cholangiocarcinoma, CORE colorectal adenocarcinoma, DLBC diffuse large B-cell lymphoma, ESCC esophageal squamous cell carcinoma, GBMLGG glioma (glioblastoma and low-grade glioma), GEAD gastric and esophageal carcinoma, HNSC head and neck squamous cell carcinoma, KIPAN pan-kidney cohort (clear cell, chromophobe, and papillary renal cell carcinoma), LAML acute myeloid leukemia, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, MESO mesothelioma, PAAD pancreatic adenocarcinoma, PCPG pheochromocytoma and paraganglioma, PRAD prostate adenocarcinoma, SARC sarcoma, SKCM skin cutaneous melanoma, TGCT testicular germ cell tumor, THCA thyroid carcinoma, THYM thymoma, UCEC uterine corpus endometrial carcinoma, UCS uterine carcinosarcoma, UVM uveal melanoma.
Since RF model prediction is based on consensus from an ensemble of individual decision trees (n = 500 for this study), the proportion of trees behind the consensus is a measure of confidence. By defining high, moderate, and low levels of prediction confidence as ≥0.90, ≥0.50 and <0.90, <0.50 of trees behind the consensus call, respectively, we showed using the smallest hybrid model that high confidence was, as expected, associated with greater accuracy (Fig. 3c). Not surprisingly, confidence levels varied across cancer reference classes (Supplementary Table 13); for example, model predictions of SCC subtypes were associated with lower confidence, possibly because carcinomas with squamous differentiation across multiple anatomic sites can have similar molecular signatures [9, 10, 37, 38].
Lastly, we assessed the impact of tumor purity on classifier accuracy and confidence for the smallest hybrid model. Tumor cellularity levels based on methylation signatures were obtained for TCGA cases from a recent publication [33]. Figure 3d shows that cases correctly classified by that model had statistically higher purities, although the magnitude of the difference between correct and incorrect cases is modest. Figure 3e shows a direct correlation between purity and confidence levels, although the strength of the association is weak (Spearman rho = 0.19).
Minimalist models can accurately classify FFPE primary and FFPE metastatic cancers
Additional minimalist classifiers were developed for classifying publicly-available and institutional datasets [3, 11,12,13,14,15,16,17,18,19,20,21] of FFPE primary and FFPE metastatic cancers. These classifiers along with the smallest hybrid TCGA classifier are summarized in Table 1. Highly-ranked probes used for the classifiers were those identified from analyses of TCGA data.
Two models were trained to classify FFPE primary cases limited to information from the same 55 probes (Supplementary Table 14; Table 1; Fig. 4a, b). Interestingly, one model, trained exclusively on TCGA fresh primary data fared poorly on validation, while the other, trained on a combination of TCGA and additional FFPE primary data performed much better—with overall accuracies of 74.6% (Fig. 4a) and 97.6% (Fig. 4b), respectively. Concordantly, the exclusively TCGA-trained model was also less confident on FFPE primary cases than the combined TCGA/FFPE-trained model.
For FFPE primary cases (a, b; n = 339, 12 cancer types), the model trained on TCGA fresh primary data (a) was far less accurate (74.6% vs. 97.6%, left panels) and less confident (right panels) on independent validation of FFPE primary cancers than the model trained on combined TCGA and randomly selected FFPE primary cases (b). Since the two models used the same 55 probes, a, b suggest that probes identified via TCGA analyses are robust—although classifiers may need to be re-trained on new data. For FFPE brain metastasis (c, d; n = 45, including three CUPs, four cancer types): c) t-SNE plot shows that FFPE brain metastasis cases (Ys, n = 42) are well-separated, and cluster with their fresh primary counterparts (dots); CUPs are indicated by black open circles. d The heat map/confusion matrix for the classifier trained on FFPE brain metastases, validated on the 39 FFPE brain metastases of known origins (six unique probes, 95.2% accuracy); CUPs are not shown here. e A single probe (cg22280705) accurately separated all nine lymph node metastases and 98.5% of 32 melanoma and 235 breast carcinoma primaries on validation. BLCA bladder carcinoma, BRCA breast invasive carcinoma, CORE colorectal adenocarcinoma, DLBC diffuse large B-cell lymphoma, GBMLGG glioma (glioblastoma and low-grade glioma), GCT germ cell tumor (intracranial), HNSC head and neck squamous cell carcinoma, KIPAN pan-kidney cohort (clear cell, chromophobe, and papillary renal cell carcinoma), LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, PRAD prostate adenocarcinoma, SKCM skin cutaneous melanoma, UCEC uterine corpus endometrial carcinoma.
A single model was trained using FFPE brain metastasis data to classify independent FFPE brain metastases, limited to information from six highly-ranked probes (Supplementary Table 15); this classifier achieved an accuracy of 95.2% (Table 1; Fig. 4c, d). t-SNE dimensionality reduction (Fig. 4c), based on information from the six CpG probes, showed that FFPE brain metastasis cases tend to cluster with their fresh primary counterparts. The CUPs included for validation were predicted to originate from the lung, identical to the original study [11].
For the lymph node metastasis dataset, we identified an optimal cutoff for a single highly-ranked probe (cg22280705, GALNT3, chr2: 166650769) from TCGA breast carcinoma versus melanoma differential; this cutoff accurately separated both TCGA fresh primary and FFPE lymph node metastases of breast cancers and melanomas on validation (Fig. 4e).
These result support the accuracy of minimalist approaches to cancer classification by methylation, at least in some formalin fixed/metastatic settings. Figure 4 suggests that retraining of classifiers on new (e.g., FFPE) data may be required.
Methylation values from a single CpG site can distinguish gastric from pancreatic adenocarcinomas
We next applied our approach to the pancreatic versus gastric adenocarcinoma differential, two cancer types that do not have sensitive and specific immunohistochemical stains. An optimal cutoff was determined for a single highly-ranked CpG probe (cg06981182, RNLS, chr10: 90343208) previously identified from TCGA analyses; Fig. 5 shows that this cutoff accurately separated independent cases of primary gastric and pancreatic adenocarcinomas from TCGA (90.0% accuracy; n = 201) and GEO [24,25,26,27,28,29,30]/ICGC (90.3%; n = 257) on validation.
The distributions of methylation values for pancreatic and gastric cancers are similar in ICGC/GEO and TCGA validation datasets. Conceivably, analysis of single CpG sites can be carried out via quantitative PCR. ICGC International Cancer Genome Consortium, GEAD gastric/esophageal adenocarcinoma, GEO Gene Expression Omnibus, PAAD pancreatic adenocarcinoma, STAD gastric adenocarcinoma.
Minimalistic cancer classifier for high-throughput methylation sequencing data
Lastly, to extend our minimalist approach to a non-array platform, we trained a 24-probe classifier (Supplementary Table 16) using TCGA and FFPE methylation array data, and tested this on 15 institutional cases profiled in parallel by methyl-seq and EPIC array (Table 1). This classifier correctly identified the tumor type with medium or high levels of confidence in 10 of 15 samples analyzed by methyl-seq (Fig. 6). The predictions and confidence levels based on methyl-seq data were generally comparable to those based on EPIC array data; concordantly, correlations of methylation values across the 24 CpG sites were high between the two platforms (Pearson R > 0.9) for 13 out of 15 cases. Interestingly, the remaining two samples with lower correlations (Cases S8, S9; Fig. 6) were correctly classified based on array data, and incorrectly classified based on methyl-seq data; we were unable to explain the lower correlations between the two platforms (via assessment of sample quality metrics) for the two cases.
We trained a classifier using data from TCGA and FFPE methylation array cases. This classifier was tested on 15 institutional FFPE primary cases, for which both methyl-seq (triangles) and EPIC array (circles) data were available (top panel). The reference cancer type is color coded in the sample name labels on the X-axis and the predictions of the classifier are color coded (by cancer type) and plotted with respect to their levels of confidence. The classifier correctly predicted 11 out of 15 diagnoses (73.3%; of which, 10 of 15 had medium or high levels of confidence) based on methyl-seq data, and 14 out of 15 diagnoses (93.3%) based on array data. Below each sample, the correlations between methylation levels across the 24 probes for the two platforms are shown (bottom panel). BLCA bladder carcinoma, CESC cervical and endocervical cancers, CORE colorectal adenocarcinoma, LUAD lung adenocarcinoma, SKCM skin cutaneous melanoma, UCEC uterine corpus endometrial carcinoma.
Gene enrichment analysis
From the top-10 ranked entries from the 378 pairwise differential lists from TCGA analysis, there were 2188 unique CpG sites, corresponding to 1176 unique genes. Using Enrichr [34, 35], we found that the genes were enriched for specific biological processes, including tissue/organ development and morphogenesis, and specific molecular functions, including cell signaling, cell-to-cell interactions, and transcriptional regulation (See Supplementary Excel Files for enriched GO biological processes, molecular functions, and cellular components), thus supporting the idea that the top-ranked diagnostic features identified from TCGA are plausible as tissue-of-origin markers.
Discussions
The potential for minimalist approaches to cancer classification by DNA methylation is supported by several considerations.
Notably, minimalist classification can be accurate (Tables 1, 2 and Figs. 3–6). As examples: the accuracy of our 53-probe pan-cancer classifiers for TCGA cases (Fig. 3) is comparable to that for published pan-cancer models, including those that employed more probes [2, 39]; the performance of our six-probe brain metastasis classifier is similar to classifiers published by Orozco et al. [11]. While minimalist classifiers generally performed well on additional fresh/FPPE primary/metastatic cases from outside TCGA [3, 11,12,13,14,15,16,17,18,19,20,21, 24,25,26,27,28,29,30], we encountered some challenges involving FFPE primary (Fig. 4) and methyl-seq cases (Fig. 6). With respect to FFPE primary cancers, our first classifier, trained exclusively on data from TCGA fresh primary samples, performed poorly on FFPE validation (Fig. 4a). This is problematic from our perspective since the majority of surgical specimens in pathology undergo formalin fixation, which alters DNA methylation patterns [40], and may contribute to loss of accuracy in methylation-based classification. We resolved this problem by training a similar model using combined TCGA and FFPE primary data limited to the same set of probes as before, and this new model performed much better (Fig. 4b). This shows that the informative probes identified from TCGA data remained robust in the FFPE primary setting, as long as the classifier was retrained on data that included FFPE cases. Conceivably, similar retraining using sequencing data could improve the accuracies and confidence levels of methyl-seq classifications, since the correlations between the platforms were often high (Fig. 6); however, we do not have sufficient methyl-seq cases at this time to train a new classifier. The methyl-seq dataset also proved problematic for a different reason: a significant proportion of CpG sites had >10% missing values across cases (see “Methods”), and we only included ~120,000 out of ~800,000 targeted CpG sites for the analysis. This means that robust methylation array biomarkers may not be consistently quantifiable via methyl-seq. Accordingly, across the various minimalist classifiers developed for this study (Table 1), general conclusions were drawn: first, TCGA datasets appear to be ideal for initial biomarker discovery, since these include large numbers of cases from diverse cancer types; second, translation of informatics analyses into robust assays for non-TCGA cases may require classifier re-training on new sample types (e.g., when going from fresh frozen to FFPE); attention to features/limitations specific to individual platforms are also required.
Further investigations along these lines may well be worthwhile, considering the potential advantages of minimalist methylation-based tests in comparison to more established methods in use today. In pathology, tissue-of-origin information is often obtained via IHC, but the development of IHC stains can be a “hit-and-miss” process. By comparison, our feature ranking studies of TCGA methylomes consistently identified large numbers of highly sensitive and specific epigenetic biomarkers for the vast majority of cancer differentials (except for those involving some types of SCCs), including many biomarkers that exceeded the performances of the best-available IHCs (Fig. 1b, c and Supplementary Tables 6–8). Consider a hypothetical case of metastatic gastric versus pancreatic adenocarcinoma; while existing IHC stains would not be helpful, our ranking analyses identified 187 probes with AUCs ≥0.9 for the pancreatic versus GEAD differential. One of these probes (cg06981182, RNLS, chr10: 90343208) is specifically highlighted in Fig. 5, to illustrate how the quantitative interrogation of a single CpG site could help resolve a potentially difficult differential.
What distinguishes minimalist approaches in our manuscript from the more common methylation classifiers based on comprehensive profiling, such as EPICUP [2] and the molecularneuropathology.org [3] model, is far smaller numbers of probe used (i.e., <0.1% of all probes on the DNA methylation array). Importantly, Fig. 2 suggests that minimalist tests could be (almost) as accurate comprehensive profiling—implying that most of the approximately 450 thousand probes on the TCGA arrays are either uninformative or informationally redundant, at least for classification in the fresh primary pan-cancer setting. This directly supports the idea that methylation-based tests can be dramatically scaled-down, without significant increases in classification errors, and thereby adapted for lower throughput and more popular platforms.
Such minimalist tests would not require array equipment, and could prove popular for smaller laboratories that have already invested, or are planning to invest, in NGS (consider Fig. 3 for a pan-cancer example) and/or quantitative PCR platforms (consider Fig. 5 for as an example involving 2 cancer types). The College of American Pathologists 2020 catalog (https://documents.cap.org/documents/2020-surveys-catalog-mobile-111119-v3.pdf) for laboratory proficiency testing highlights surveys available for NGS- and PCR-based platforms, but does not include a survey specific for methylation array profiling—reflecting the relative rarity of the latter in the present-day “clinical space.” With respect to NGS testing, potential advantages of targeting small numbers of CpG sites also include higher sequencing depths and greater multiplexing; the higher depths may produce better data for some CpG sites in comparison to methyl-seq, which is characterized by higher sequencing breadth but less depth at individual loci. Further, for quantitative PCR, one anticipated advantage is lower costs per sample, at least for limited differentials. For reference, the reagent cost of methylation array profiling is ~500 Canadian dollars (CAD) per sample at one of our research laboratories. We estimate that quantitative PCRs could be significantly less expensive, at ~10–15 CAD for reagents per reaction/CpG site (see in Fig. 5 as a hypothetical example). However, it should be cautioned that actual costs and implementation depend on samples per run and overall volume, and CUPs and other difficult cases are only small fractions of clinical volumes on most pathology services.
The final and related consideration is that while poor quality formalin-fixed and bisulfite converted DNA can be challenging to work with, lower throughput technologies for the quantitative interrogation of small numbers of CpG sites in FFPE samples already exist [11, 41, 42]. For instance, Orozco et al. [11] described quantitative methylation-specific PCR assays for distinguishing (1) gliomas from brain metastases, (2) lung, breast, and melanoma brain metastases from one another, and (3) HER2+ from HER2− breast cancers metastatic to the brain. Van Wesenbeeck et al. [41] developed a droplet digital PCR protocol for the quantitative assessment of two CpG sites on FFPE samples of colonic adenomas. Importantly, correlations between methylation levels measured by arrays and PCRs were high [41], supporting the possibility of robust quantitative testing across multiple platforms—once the most informative biomarkers are identified. We acknowledge the value of the relatively few published minimalist tests for specific cancer types/applications, and note that our work on generalized strategies suggest that similar efforts can be applied to a much broader array of challenging differentials in pathology.
In terms of limitations, the scope of this study and its results/conclusions are limited to the types of cancers and samples included. For additional validation, we attempted to include diverse fresh/FFPE primary and metastasis cases mostly from outside TCGA (Figs. 4–6: n = 665 cases across 16 cancer types from 20 publicly-available and two internal datasets). While this supports the general robustness of minimalist approaches, our combined data is limited: there were few CUPs, relatively few metastases, few cases of some cancer types, no cases of some potentially important cancer types, and few cases based on non-array data. Our proof-of-principle study is therefore not intended to recapitulate minimalist testing on CUPs, or to address every conceivable difficult differential across multiple testing platforms. Further, cancers with low tumor purities, which are not uncommon in clinical practice, are likely underrepresented because, all else being equal, those with higher purities are usually preferentially selected for profiling. While Fig. 3d, e shows modest correlations between tumor purity and classifier confidence and accuracy, this applies only to TCGA cases.
On balance, a compelling case in support of further investigations into minimalist approaches to cancer classification by DNA methylation can be advanced, based on the generally excellent diagnostic performances of minimalist classifiers, the present availability of testing platforms, and considerations pertaining the practical needs and resources of individual (particularly smaller) laboratories. Consider an analogy to somatic mutation testing: while comprehensive whole genome/exome sequencing defined the reference mutational landscapes of cancers, clinical testing today is dominated by single gene tests and targeted NGS panels, in attempts to balance clinical needs and practical consideration against exploratory research potential (Table 3). We propose that future efforts could focus on the development of minimalist testing on wide-available platforms for CUPs and additional strategically-selected differentials in pathology subspecialties, particularly for differentials not reliably classified via traditional methods such as IHCs. Inclusion of low purity specimens is also advised to help define limits of detections and analytical sensitivities of assays.
Data availability
Data processing and analyses were completed using R (version 3.2.3) [36]. Processed data files, R scripts, lists of top-ranked CpG features, and the results of analyses not included with this manuscript may be available from the authors upon request, but could be subject to Material/Data Transfer Agreement(s).
References
Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet. 2012;379:1428–35.
Moran S, Martínez-Cardús A, Sayols S, Musulén E, Balañá C, Estival-Gonzalez A, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17:1386–95.
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–74.
Economopoulou P, Mountzios G, Pavlidis N, Pentheroudakis G. Cancer of unknown primary origin in the genomic era: elucidating the dark box of cancer. Cancer Treat Rev. 2015;41:598–604.
Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–44.
Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e6.
Mamatjan Y, Agnihotri S, Goldenberg A, Tonge P, Mansouri S, Zadeh G, et al. Molecular signatures for tumor classification: an analysis of the cancer genome atlas data. J Mol Diagn. 2017;19:881–91.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Cancer Genome Atlas Research Network. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–75.
Peyser ND, Grandis JR. Cancer genomics: spot the difference. Nature. 2017;541:162–3.
Orozco JIJ, Knijnenburg TA, Manughian-Peter AO, Salomon MP, Barkhoudarian G, Jalas JR, et al. Epigenetic profiling for the molecular classification of metastatic brain tumors. Nat Commun. 2018;9. https://doi.org/10.1038/s41467-018-06715-y.
Salomon MP, Orozco JIJ, Wilmott JS, Hothi P, Manughian-Peter AO, Cobbs CS, et al. Brain metastasis DNA methylomes, a novel resource for the identification of biological and clinical features. Sci Data. 2018;5:180245
Fukushima S, Yamashita S, Kobayashi H, Takami H, Fukuoka K, Nakamura T, et al. Genome-wide methylation profiles in primary intracranial germ cell tumors indicate a primordial germ cell origin for germinomas. Acta Neuropathol. 2017;133:445–62.
Marzese DM, Scolyer RA, Huynh JL, Huang SK, Hirose H, Chong KK, et al. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox d cluster associated with prognosis. Hum Mol Genet. 2014;23:226–38.
De Ruijter TC, De Hoon JP, Slaats J, De Vries B, Janssen MJ, Van Wezel T, et al. Formalin-fixed, paraffin-embedded (FFPE) tissue epigenomics using Infinium HumanMethylation450 BeadChip assays. Lab Investig. 2015;95:833–42.
Mathe A, Wong-Brown M, Locke WJ, Stirzaker C, Braye SG, Forbes JF, et al. DNA methylation profile of triple negative breast cancer-specific genes comparing lymph node positive patients to lymph node negative patients. Sci Rep. 2016;6:1–15.
Jour G, Vasudevaraja V, Prieto VG, Snuderl M, Torres-Cabala CA, Al-Rohil R, et al. BCAT1 and miR-2504: novel methylome signature distinguishes spindle/desmoplastic melanoma from superficial malignant peripheral nerve sheath tumor. Mod Pathol. 2019;32:338–45.
Conway K, Edmiston SN, Parker JS, Kuan PF, Tsai YH, Groben PA, et al. Identification of a robust methylation classifier for cutaneous melanoma diagnosis. J Investig Dermatol. 2019;139:1349–61.
Bady P, Kurscheid S, Delorenzi M, Gorlia T, van den Bent MJ, Hoang-Xuan K, et al. The DNA methylome of DDR genes and benefit from RT or TMZ in IDH mutant low-grade glioma treated in EORTC 22033. Acta Neuropathol. 2018;135:601–15.
Flower KJ, Shenker NS, El-Bahrawy M, Goldgar DE, Parsons MT, Spurdle AB, et al. DNA methylation profiling to assess pathogenicity of BRCA1 unclassified variants in breast cancer. Epigenetics. 2015;10:1121–32.
Lechner M, Fenton T, West J, Wilson G, Feber A, Henderson S, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med. 2013;5:1–16.
Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462
Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl J Med. 2015;372:2481–98.
Kwon OH, Park JL, Kim M, Kim JH, Lee HC, Kim HJ, et al. Aberrant up-regulation of LAMB3 and LAMC2 by promoter demethylation in gastric cancer. Biochem Biophys Res Commun. 2011;406:539–45.
Zouridis H, Deng N, Ivanova T, Zhu Y, Wong B, Huang D, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med. 2012;4. https://doi.org/10.1126/scitranslmed.3004504.
Lei Z, Tan IB, Das K, Deng N, Zouridis H, Pattison S, et al. Identification of molecular subtypes of gastric cancer with different responses to pi3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–65.
Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–97.
Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Nones K, Waddell N, Song S, Patch AM, Miller D, Johns A, et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int J Cancer. 2014;135:1110–8.
Timp W, Bravo HC, McDonald OG, Goggins M, Umbricht C, Zeiger M, et al. Large hypomethylated blocks as a universal defining epigenetic alteration in human solid tumors. Genome Med. 2014;6:1–11.
Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11. https://doi.org/10.1186/1471-2105-11-587.
Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017;45:e22.
Zheng X, Zhang N, Wu HJ, Wu H. Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies. Genome Biol. 2017;18:1–14.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7.
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128
R Core Team. R: a language and environment for statistical computing. 2013. http://www.r-project.org/.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82.
Tang W, Wan S, Yang Z, Teschendorff AE, Zou Q. Tumor origin detection with tissue-specific miRNA and DNA methylation markers. Bioinformatics. 2018;34:398–406.
Kling T, Wenger A, Beck S, Carén H. Validation of the MethylationEPIC BeadChip for fresh-frozen and formalin-fixed paraffin-embedded tumours. Clin Epigenetics. 2017;9:1–6.
Van Wesenbeeck L, Janssens L, Meeuws H, Lagatie O, Stuyver L. Droplet digital PCR is an accurate method to assess methylation status on FFPE samples. Epigenetics. 2018;13:207–13.
Giacopelli B, Zhao Q, Ruppert AS, Agyeman A, Weigel C, Wu YZ, et al. Developmental subtypes assessed by DNA methylation-iPLEX forecast the natural history of chronic lymphocytic leukemia. Blood. 2019;134:688–98.
Acknowledgements
This study was conducted with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario, via the 2018–19 Ontario Molecular Pathology Research Network Cancer Pathology Translational Research Grant (#CPTRG-016), awarded to DX and KA. The internal lung cancer methylation profiling was supported by the Canadian Institute of Health Research (CIHR) Foundation Grant (FDN-148395) and Lung Cancer Research Fund of the Princess Margaret Hospital (PMH) Foundation. MC is supported by the Terry Fox Foundation Training Program in Molecular Pathology of Cancer at CIHR (STP 53912). FAS is the Scott Taylor Chair in Lung Cancer Research. MST is the M. Qasim Choksi Chair in Lung Cancer Translational Research. We thank the staff of the Translational Genomics Laboratory (https://labs.oicr.on.ca/translational-genomics-laboratory) for their expertise in generating and analyzing the sequencing data used in this study. The Translational Genomics Laboratory is a joint initiative between the Princess Margaret Cancer Center and the Ontario Institute for Cancer Research that is enabled through funding provided by the Government of Ontario, and the Princess Margaret Cancer Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
DX and AJL are listed as contributors in a submitted provisional patent application. Other authors have no potential conflict of interest to declare.
Ethical approval
The protocols relevant to this study were approved by the Research Ethics Board at the University Health Network (UHN; CAPCR IDs 18–5465, 13–6879, and 12–5453).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xia, D., Leon, A.J., Cabanero, M. et al. Minimalist approaches to cancer tissue-of-origin classification by DNA methylation. Mod Pathol 33, 1874–1888 (2020). https://doi.org/10.1038/s41379-020-0547-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-020-0547-7
This article is cited by
-
Comprehensive analysis of cancer of unknown primary and recommendation of a histological and immunohistochemical diagnostic strategy from China
BMC Cancer (2023)
-
Hierarchical classification-based pan-cancer methylation analysis to classify primary cancer
BMC Bioinformatics (2023)
-
DICER1-sarcoma: an emerging entity
Modern Pathology (2021)
-
Histologische, immunhistologische und molekularpathologische Aspekte bei nichtmelanozytären Hauttumoren
Der Onkologe (2021)