Abstract
There is now evidence that gene fusions activating the MAPK pathway are relatively common in pancreatic acinar cell carcinoma with potentially actionable BRAF or RET fusions being found in ~30%. We sought to investigate the incidence of RAF1 fusions in pancreatic malignancies with acinar cell differentiation. FISH testing for RAF1 was undertaken on 30 tumors comprising 25 ‘pure’ acinar cell carcinomas, 2 mixed pancreatic acinar-neuroendocrine carcinomas, 1 mixed acinar cell-low grade neuroendocrine tumor and 2 pancreatoblastomas. RAF1 rearrangements were identified in 5 cases and confirmed by DNA and RNA sequencing to represent oncogenic fusions (GATM-RAF1, GOLGA4-RAF1, PDZRN3-RAF1, HERPUD1-RAF1 and TRIM33-RAF1) and to be mutually exclusive with BRAF and RET fusions, as well as KRAS mutations. Large genome-wide copy number changes were common and included 1q gain and/or 1p loss in all five RAF1 FISH-positive acinar cell carcinomas. RAF1 expression by immunohistochemistry was found in 3 of 5 (60%) of fusion-positive cases and no FISH-negative cases. Phospho-ERK1/2 expression was found in 4 of 5 RAF1-fusion-positive cases. Expression of both RAF1 and phospho-ERK1/2 was heterogeneous and often only detected at the tumor-stroma interface, thus limiting their clinical utility. We conclude that RAF1 gene rearrangements are relatively common in pancreatic acinar cell carcinomas (14.3% to 18.5% of cases) and can be effectively identified by FISH with follow up molecular testing. The combined results of several studies now indicate that BRAF, RET or RAF1 fusions occur in between one third and one-half of these tumors but are extremely rare in other pancreatic malignancies. As these fusions are potentially actionable with currently available therapies, a strong argument can be made to perform FISH or molecular testing on all pancreatic acinar cell carcinomas.
Similar content being viewed by others
Introduction
Pancreatic acinar cell carcinoma is an uncommon form of pancreatic neoplasia. It accounts for only 1–2% of all pancreatic carcinomas in adults, but is relatively more common in children (up to 15% of pediatric pancreatic neoplasms) where there is some morphological and clinical overlap with pancreatoblastoma from which it is differentiated by the presence of squamous morules [1,2,3]. In contrast to pancreatic ductal adenocarcinoma which is associated with somatic KRAS mutations in more than 93% of cases [4,5,6], mutations in KRAS are very rare in acinar cell carcinoma and mutations in the other most common pancreatic ductal adenocarcinoma driver genes including TP53, CDKN2A, and SMAD4 are infrequent [7,8,9]. Importantly the combined results of several recent studies have demonstrated that potentially targetable activating gene rearrangements in BRAF and RET that activate the MAPK signaling pathway are relatively common and may be the driver event in more than 30% of acinar cell carcinomas [10,11,12,13,14].
A single patient with a RAF1 rearrangement was reported previously in a study of 44 pancreatic acinar cell carcinomas screened by panel DNA sequencing [10]. We also recently reported a 33-year-old male with pancreatic acinar cell carcinoma associated with a GATM-RAF1 fusion oncogene identified by a personalized medicine approach [15]. RAF1 fusions are significant as they are targetable with already available agents (e.g., MEK inhibitors and RAF dimerization inhibitors) and significant responses to MEK inhibition have been reported in melanoma and anaplastic pleomorphic xanthoastrocytoma associated with RAF1 gene rearrangements [16,17,18].
Given the high incidences of gene rearrangements in BRAF and RET in pancreatic acinar cell carcinoma, we postulated that RAF1 gene rearrangements (which also activate the MAPK signaling pathway) may be common in this malignancy. We, therefore, sought to study the incidence of RAF1 gene rearrangements in a large cohort of acinar cell carcinomas and investigate whether these rearrangements are mutually exclusive with the previously reported BRAF and RET rearrangements. Furthermore, we performed immunohistochemistry for pERK to determine if RAF1 rearrangements, when present, activated the MAPK pathway.
Methods
We searched the institutional databases of multiple centers for all pancreatic malignancies reported as demonstrating some acinar cell differentiation. We included both pure pancreatic acinar cell carcinomas and mixed pancreatic acinar cell carcinoma–neuroendocrine tumors – defined in accordance with the WHO 2019 criteria as tumors demonstrating greater than 30% of both acinar and neuroendocrine differentiation [2]. For the purposes of this study, pancreatoblastomas, defined in accordance with the WHO 2019 system as tumors with both acinar differentiation and morphologically identified squamous morules were also included [2].
All cases demonstrated unequivocal acinar cell differentiation (by both morphology and positive immunohistochemical staining for both BCL10 and trypsin) and underwent centralized pathological review by at least two surgical pathologists with expertize in pancreatic pathology to confirm the diagnosis. Inclusion criteria required sufficient tumor in formalin-fixed paraffin-embedded (FFPE) blocks for further testing. Clinicopathological data collected included tumor size, age, sex, date of surgery, type of operation, type of sample, size of tumor, metastasis at presentation, lymph node involvement and AJCC pathological stage.
Fluorescent in-situ hybridization (FISH) was performed on FFPE sections using a RAF1 dual-color break-apart probe (Empire Genomics, New York, USA). The methods have been previously described in detail [18], briefly, the RAF1 probe has fluorescence-labeled probes that hybridize to genomic sequences that are 5’ (green) and 3’ (red) to the RAF1 gene which is located at the 3p25.2 chromosomal region. Rearrangement-positive tumor cells were defined as the presence of either a split green and red signal at least two signals diameter apart or a fused green/red signal together with an isolated red (3’) signal. Rearrangement-negative tumor cells were defined by the detection of only fused green and red signals. At least 50 tumor nuclei were counted, and for a case to be considered as RAF1 FISH-positive, at least 15% of the tumor nuclei were required to demonstrate rearranged signals. The RET and BRAF gene rearrangement status determined by FISH have been previously reported for the majority of these cases [13, 14].
DNA sequence analysis of the coding regions of 386 genes implicated in cancer (2.34 Mb in total) was performed on the RAF1 FISH-positive cases. Full methods have been previously described [19]. This panel includes the entire coding sequences of many receptor tyrosine kinases (RET, ALK, ROS1, NTRK1, NTRK3, FGFR1, FGFR2, FGFR3, PDGFB, KIT) and many MAPK pathway genes (including RAF1, BRAF, NRAS, KRAS, HRAS, GNAS, GNAQ, MAP2K1/2, MAPK1/2, NF1 and NF2) and intron coverage for common break points in RAF1 (introns 7-11), BRAF (introns 4-9), RET, ALK, ROS1, NTRK1, NTRK3, FGFR1, FGFR2, FGFR3 and PDGFB. Fusions were detected using a multithreaded structural variant (SV) calling platform - Genome Rearrangement IDentification Software Suite (GRIDSS) [20].
One patient had incidentally been enrolled in a clinical trial designed to identify predictive therapies in rare cancer types based on genomic changes identified by whole-genome sequencing (WGS). Therefore, in contrast to the other 4 RAF1-rearranged cases, only WGS and not panel DNA sequencing was performed on this case. For WGS library preparation of genomic DNA from tumor and normal samples was performed with the Illumina TruSeq Nano DNA library preparation kit following manufacturer’s instructions. In brief, whole-genome libraries were normalized, pooled and sequenced by Illumina NovaSeq 6000, and data analyzed with Illumina software Sequencing Analysis Viewer using the default Illumina pipeline. Structural variants were called with Manta SV caller [21], filtered and annotated with SnpEff [22] based on Ensembl gene models. Annotations were subset to APPRIS principal transcripts. Structural variants, including gene fusions were prioritized using simple_sv_annotation [23].
Fusions were confirmed by reverse transcription-polymerase chain reaction (RT-PCR/Sanger sequencing, cases 9, 12, 17, and 30) and transcriptome (RNA) sequencing (cases 9, 17, 29 and 30). For RNA sequencing ribosomal RNA depletion was performed by either Illumina TruSeq stranded total RNA Gold kit following manufacturer’s instructions or NEBNext Ultra II Directional RNA Library Prep kit with a modified protocol to include an on-bead PCR [19]. Whole transcriptome libraries were sequenced as described above for whole-genome libraries. RNA reads were aligned using STAR [24]. Fusions were called with Arriba and Jaffa.
Immunohistochemistry (IHC) and immunofluorescence were performed with mouse monoclonal anti-RAF1 (E-10, Santa Cruz Biotechnology INC. sc-7267) and rabbit monoclonal anti-phospho-ERK (ERK1/2 Thr202/Tyr204, Cell Signaling, #4370) – full methods previously described [18]. The same anti-RAF1 antibody was used for both IHC and immunofluorescence. For immunofluorescence the Perkin Elmer 4-Color Manual Opal kit was used according to manufacturer’s instructions and imaged with a Vectra multispectral microscope. Continuous variables were compared using Student’s t test and categorical variables were compared using Fisher’s exact test. A p value of <0.05 was considered statistically significant. The study was approved by the Northern Sydney Local Health District Human Research Ethics Committee – ref: LNR 1312-417M.
Results
A total of 30 cases of pancreatic carcinomas demonstrating acinar differentiation with sufficient material for testing were identified. These comprised 25 ‘pure’ pancreatic acinar cell carcinomas (ACCs, n = 25), 2 mixed pancreatic acinar cell carcinoma–neuroendocrine carcinomas (mixed ACC-NEC, n = 2), 1 mixed pancreatic acinar cell carcinoma–low grade neuroendocrine tumor (mixed ACC-NET, n = 1) and 2 pancreatoblastomas (PB, n = 2). These 30 cases included the index acinar cell carcinoma with a GATM-RAF1 fusion which we had previously reported (index case, patient 12) [15]. The clinicopathological characteristics of the cohort and the results of FISH testing are summarized in Table 1. There were 23 males and 7 females with a median age of 62 years (mean 57 years, range 17–82 years). The median tumor size was 39 mm (mean 51 mm, range 13–190 mm). 8 patients presented with stage I disease, 10 with stage II disease, 1 with stage III disease and 10 with stage IV disease. The stage was unknown for 1 patient. The index case with the GATM-RAF1 fusion (patient 12) was confirmed by break-apart RAF1 FISH, with loss of the 5’ signal. Break-apart RAF1 FISH was successful in 28 of 29 of the other acinar cell carcinomas. FISH failed repeatedly in one patient due to high green background intensity. Of the 28 assessable new cases, four demonstrated RAF1 FISH patterns consistent with gene rearrangement (Fig. 1). Two cases showed loss of the 5’ (green) signal and two cases showed split 5’/3’ signals (Table 1). These rearranged FISH patterns were present throughout the neoplastic cells within the tumors without evidence of heterogeneity.
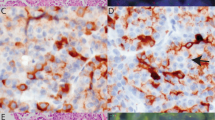
RAF1-rearranged acinar cell carcinoma case 9 (a H&E morphology; b RAF1 break-apart FISH, arrows showing split 5’/3’ signals; c RAF1 immunostains showing granular cytoplasmic staining, inset is RAF1 immunofluorescence [red] and nuclei [aqua]). RAF1-rearranged acinar cell carcinoma case 30 (d H&E morphology; e RAF1 break-apart FISH, arrows showing isolated 3’ [red] signals; f RAF1 immunostains showing granular cytoplasmic staining). RAF1-negative acinar cell carcinoma case 5 (g H&E morphology; h RAF1 break-apart FISH showing fused 5’/3’ signals; i RAF1 immunostains showing weak diffuse cytoplasmic staining in carcinoma and strong diffuse cytoplasmic staining in benign islet cells [arrowhead]). Scale bar 100 µm in a, d, g; 50 µm in c, f, i.
The clinicopathological characteristics of all five RAF1 FISH-positive cases (that is the index case plus the further four positive cases) are summarized in Table 1. The histological subtype was either pure acinar cell carcinoma (4 cases) or mixed acinar cell carcinoma–neuroendocrine carcinoma (1 case). Both cases of pancreatoblastoma were RAF1 FISH-negative. The overall RAF1 FISH-positive incidence was therefore 5 of 29 cases (17.2%), and 5 of 27 (18.5%) if pancreatoblastomas were excluded. The RAF1 FISH-positive incidence in the unselected cases (i.e., excluding the index case) was 4 of 28 (14.3%), and 4 of 26 (15.4%) if pancreatoblastomas were excluded. Univariate analysis demonstrated no significant difference between tumors with and without RAF1 arrangement (Table 2) based on gender (p = 1.000), age at presentation (p = 0.637), size (p = 0.600), metastasis at presentation (p = 0.306), nodal involvement at presentation (p = 1.000) and pathological stage (p = 0.430).
In addition to the index case which harbored a GATM-RAF1 fusion, RAF1 fusions were confirmed in all four additional RAF1 FISH-positive cases by DNA sequencing - summarized in Table 3 and Fig. 2. The fusions were GOLGA4-RAF1, PDZRN3-RAF1, HERPUD1-RAF1 and TRIM33-RAF1. Two of the 5’ fusion partners (GOLGA4 and PDZRN3) were on the same chromosomal arm as RAF1 (3p). The gene orientation of GOLGA4 and RAF1, split 5’/3’ RAF1 FISH pattern, and detection of both GOLGA4-RAF1 and RAF1-GOLGA4 fusion transcripts suggested an intrachromosomal inversion. In-frame, expressed RAF1 fusion transcripts were detected by RNA sequencing in all cases (Table 3). These all removed the N-terminal RAS-binding (RBD) and C1 auto-inhibitory domains and maintained an intact RAF1 kinase domain, likely contributing to increased kinase activity [25]. Interestingly, DNA sequencing of the acinar cell carcinoma from patient 29 demonstrated that RAF1 exons 8-17 were fused to the 3’ untranslated region of HERPUD1, which was not predicted to yield a functional fusion protein. However, RNA sequencing detected low frequency in-frame chimeric transcripts with HERPUD1 exons 1-7 fused to RAF1 exons 8-17, consistent with skipping of HERPUD1 exon 8 and its stop codon. This tumor also harbored an activating missense mutation in MEK1 (MAP2K1). Aside from the RAF1 fusions, mutations in the MAPK pathway were not detected in the other four acinar cell carcinomas. The index case with the GATM-RAF1 fusion was surprisingly both RAF1 and BRAF FISH-positive, but there was no evidence of a BRAF structural variant by DNA sequencing. Where tested, none of the other RAF1 FISH-positive acinar cell carcinomas had BRAF or RET rearrangements by FISH (Table 1).
Five RAF1 fusions were identified by DNA and RNA sequencing. The chromosomal (Chr) locations of the fusion partners for these are shown, together with the number of mapped paired reads, the involved exons (Ex), and the protein domains predicted in the chimeric proteins (MTP: Mitochondrial transit peptide; RING: RING finger UBL: Ubiquitin-like; RBCC: Ring, B-box, Coiled-Coil).
Large genome-wide copy number changes were common and included 1q gain and/or 1p loss in all five RAF1 FISH-positive acinar cell carcinomas. Similar changes have been reported in acinar cell carcinomas by others [8]. Similar to pancreatic ductal adenocarcinoma there was often homozygous deletion of either or both CDKN2A and CDKN2B (3 of 5 tumors) and inactivating mutations in SMAD4 (2 of 5 tumors). However, in distinction to pancreatic ductal adenocarcinoma all of the RAF1 FISH-positive acinar cell carcinomas were wild-type for KRAS and TP53.
IHC for RAF1 with an antibody directed to an epitope between amino acids 637-648 at the C-terminus of human RAF-1 was performed on all 29 cases with a RAF1 FISH result. RAF1 IHC was successful in 25 cases (5 RAF1 FISH-positive, 20 RAF1 FISH-negative) as evidenced by weak fine diffuse cytoplasmic expression in pancreatic islets, benign acinar cells, endothelial cells and inflammatory cells, a pattern similar to that reported in the Human Protein Atlas [26]. Positive RAF1 IHC, as defined by at least focally substantially more intense staining in tumor cells than in benign cells, was detected in 3 of 5 RAF1 FISH-positive acinar cell carcinomas and 0/20 RAF1 FISH-negative acinar cell carcinomas (Table 3). The 3 RAF1 IHC-positive cases (patients 9, 12, 30) all showed moderate to strong granular cytoplasmic staining (Figs. 1 and 3), a pattern that was confirmed by immunofluorescence (Fig. 1c). However, RAF1 staining was highly heterogeneous. It was most intense at tumor-stroma interfaces and in acini (Figs. 1 and 3), but many of the tumors displayed large areas with negative staining.
RAF1 expression in case 9 was enhanced at the tumor-stroma interface (a, main image) and in acini (a, inset), and at the tumor-stroma interface of case 12 (c). There was only weak diffuse cytoplasmic RAF1 staining in tumor cells in cases 17 (e) and 29 (g) that was less intense than in endothelial cells. pERK expression was intense at the tumor-stroma interface in case 9 (b, main image and inset), focally at the tumor-stroma interface in case 17 (f), and throughout the tumor in cases 12 (d) and 29 (h). Scale bar 200 µm in main images a, b, 100 µm in e, f, 50 µm in insets a, b and in c, d, g, h.
Signaling downstream of RAF1 targeted to the plasma membrane typically activates the MAPK pathway resulting in phosphorylation and activation of ERK1/2 (p44/42 MAPK). IHC for phospho-ERK1/2 (pERK) was performed on all 29 cases with a RAF1 FISH result and was interpretable in 20 cases as evidenced by cytoplasmic and nuclear expression in internal controls (intra-tumoral endothelial cells). pERK staining was detected in the tumor cells of 4/5 RAF1 FISH-positive tumors (cases 9, 12, 17 and 29; Fig. 3 and Table 3), one BRAF FISH-positive/RAF1 FISH-negative tumor (case 28), and in 6/12 RAF1/BRAF/RET FISH-negative tumors (cases 2, 8, 10, 21, 15, 20). pERK expression was often strongest at the tumor-stroma interface in both fusion-positive and negative cases. Notably, pERK staining was strong throughout the tumor in case 29 with the HERPUD1-RAF1 fusion and the activating MEK1 mutation.
Discussion
Although a single pancreatic acinar cell carcinoma with a RAF1 rearrangement was previously identified by large panel DNA sequencing in a cohort of 44 cases [10], and we recently identified a second RAF1-rearranged case [15], this is the first study to screen a large cohort of pancreatic tumors with acinar cell differentiation specifically by RAF1 FISH. The incidence of RAF1 fusions in this cohort was 14.3% in all unselected (excluding the index) cases. It is possible that the incidence of RAF1 fusions in acinar cell carcinoma may be greater as this figure rose to 18.5% when the cohort included the index case and excluded the 2 pancreatoblastomas. This incidence is perhaps surprising as only a single RAF1 fusion (HACL1-RAF1) was previously reported in a series of 44 acinar carcinomas [10]. Factors that may contribute to this apparent discrepancy include the relatively small cohort sizes in both studies, and the different sequencing and bioinformatic methodologies.
BRAF gene rearrangements have been reported in 14–20% of acinar cell carcinomas [10,11,12,13], and RET gene rearrangements in 8% [14]. Therefore, the combined results of this and other studies indicate that rearrangements involving the MAPK pathway genes BRAF, RET or RAF1 occur in between one third and one half of all acinar cell carcinomas. It is worth noting that ATG7-RAF1 and PDZRN3-RAF1 fusions have been reported in 3 pancreatic ductal adenocarcinomas 2 of which were documented as KRAS wild-type [27, 28]. We have reviewed the data from the Australian Pancreatic Genome Initiative (APGI) cohort of 456 pancreatic carcinomas [4,5,6] which included only two patients with acinar cell carcinoma (and one more reclassified as acinar cell carcinoma on review only after a RET gene rearrangement was identified) [13] and note that no RAF1 rearrangements were identified in this cohort consisting almost entirely of pancreatic ductal adenocarcinomas. We therefore postulate that many if not all of these previously reported pancreatic ductal adenocarcinomas with RAF1 rearrangement may in fact represent misclassified acinar cell carcinomas. Ultimately, only systematic screening of larger cohorts of bona fide acinar carcinomas and complete morphological review of pancreatic malignancies with fusions will be required to definitively determine whether RAF1 fusions are limited to acinar cell carcinomas or also occur in pancreatic ductal adenocarcinomas. In any case, regardless of whether or not these fusions occur (extremely rarely) in pancreatic ductal adenocarcinoma, this study provides unequivocal evidence that RAF1 rearrangements are markedly over-represented in acinar cell carcinomas compared to other pancreatic malignancies.
Based on current data, it appears that gene rearrangements involving any one of BRAF, RET and RAF1 in pancreatic acinar cell carcinoma are almost always mutually exclusive with each other and with KRAS mutation, a key driver event in the overwhelming majority (more than 93%) of non-acinar pancreatic ductal adenocarcinomas [4,5,6]. This mutual exclusivity of key driver events in the MAPK signaling pathway is similar to that previously reported in a large a pan-cancer cohort of solid tumors (n = 4871) [29]. This was true in all but one acinar cell carcinoma in our cohort which harbored a HERPUD1-RAF1 fusion and an activating MEK1 mutation (E203K). The HERPUD1-RAF1 RNA was expressed at very low levels, requiring skipping of HERPUD1 exon 8, and RAF1 protein was undetectable by immunohistochemistry. Therefore, it is possible that MAPK pathway activation in this carcinoma required MEK1 mutation in addition to the HERPUD1-RAF1 fusion. Consistent with this interpretation, MEK1 E203K drives ERK signaling (‘activator’ mutation) but can be further activated in a RAS/RAF-dependent matter (‘amplifier’ mutation) [30]. This suggests that broad molecular sequencing is an important orthogonal confirmation of FISH and may be critical to direct targeted therapy, as anti-RAF1 therapy alone may fail in this patient because of downstream MEK1 activation.
Several different techniques are used in clinical practice to identify gene rearrangements. FISH has the advantages of widespread availability, rapid turnaround time, minimal cost and low failure rate - 1 of 30 (3.3%) in our series. It is reassuring that in this study all 5 cases with RAF1 gene rearrangements identified by FISH were confirmed to harbor fusions by orthogonal molecular testing with both DNA and RNA sequencing indicating that FISH testing is highly specific. Whilst we did not investigate the sensitivity of FISH in this study by performing DNA and RNA sequencing on FISH-negative cases, it is likely that FISH identifies the overwhelming majority of RAF1-rearranged cases given its low incidence in previous studies [10]. However FISH has several disadvantages compared to other techniques – most importantly that it does not identify the partner genes (GATM-RAF1, GOLGA4-RAF1, PDZRN3-RAF1, HERPUD1-RAF1 and TRIM33-RAF1 in this study) and may therefore identify rearrangements that reflect chromosomal instability (common in acinar cell carcinoma [8, 10] and not always associated with pathogenic fusions). For example, in this study we identified one case that was both BRAF and RAF1 FISH-positive but was found on follow up DNA and RNA sequencing to harbor only a RAF1 and not a BRAF fusion. It is likely in this case that the BRAF FISH result was effectively a false positive due to the broad chromosomal changes that we and others [8, 10] have observed in acinar cell carcinoma. In any case, whether or not RAF1 FISH is validated as a sensitive and clinically useful screening test in other cohorts, it highlights the clear additional benefit of performing confirmatory orthogonal molecular validation of rearranged cases when resources allow.
RAF1 immunohistochemistry was positive in 3/5 RAF1-rearranged acinar carcinomas, and negative in 20/20 RAF1 FISH-negative cases. pERK immunohistochemistry was positive in 5/6 RAF1/BRAF-rearranged cases providing evidence that the fusions were functional and the MAPK pathway was activated. Overall, 6/6 (100%) RAF1 or BRAF fusion cases showed expression of either RAF1 or pERK, suggesting the potential for screening for these fusion by IHC. However, currently we would not recommend these surrogate immunohistochemical markers in routine clinical practice. Both RAF1 and pERK expression were most intense and often only detectable at the tumor-stroma interface. We noted a similar pattern in a melanoma with a GOLGA4-RAF1 fusion [18], possibly due to the microenvironment at the invasive front and subcellular compartmentalization. The staining for both RAF1 and pERK was typically absent in tumor cells away from the invasive front, and this may produce a negative result in small biopsies. The signal intensities for both also deteriorated relatively rapidly following sectioning. This unfortunately suggests that RAF1 and pERK IHC using current techniques and antibodies are not sufficiently sensitive to screen for RAF1 (or BRAF and RET) rearrangements. pERK was also detectable in 6/12 FISH-negative cases. Although we did not perform sequencing on these cases and cannot completely exclude the possibility of a fusion in RAF1, BRAF or RET that was undetected by breakapart FISH, it suggests the presence of other activating mutations in the MAPK pathway in these acinar cell carcinomas.
In conclusion, depending on whether or not pancreatoblastomas and the index case are included to limit selection bias, the incidence of RAF1 rearrangements ranges in pancreatic acinar cell carcinomas ranges from 14.3% to 18.5% in our study. Furthermore, combined with the results of other studies, one of RAF1, BRAF or RET rearrangements appear to be found in between one third and one half of all pancreatic acinar cell carcinomas. We therefore recommend a low threshold for considering the diagnosis of acinar cell carcinoma and, in confirmed cases of acinar cell carcinoma, proceeding to molecular testing to detect potentially clinically significant gene rearrangements.
References
Schmidt CM, Matos JM, Bentrem DJ, Talamonti MS, Lillemoe KD, Bilimoria KY. Acinar cell carcinoma of the pancreas in the United States: prognostic factors and comparison to ductal adenocarcinoma. J Gastrointest Surg. 2008;12:2078–86.
La Rosa S, Klimstra DS, Wood LD. Pancreatic acinar cell carcinoma. In: WHO classification of tumours: digestive system tumours. 5th ed. Lyon: IARC Press; 2019. pp 333–336.
Klimstra DS, Adsay V. Acinar neoplasms of the pancreas-A summary of 25 years of research. Semin Diagn Pathol. 2016;33:307–18.
Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405.
Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501.
Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Furukawa T, Sakamoto H, Takeuchi S, Ameri M, Kuboki Y, Yamamoto T, et al. Whole exome sequencing reveals recurrent mutations in BRCA2 and FAT genes in acinar cell carcinomas of the pancreas. Sci Rep. 2015;5:8829.
Jakel C, Bergmann F, Toth R, Assenov Y, van der Duin D, Strobel O, et al. Genome-wide genetic and epigenetic analyses of pancreatic acinar cell carcinomas reveal aberrations in genome stability. Nat Commun. 2017;8:1323.
Bergmann F, Aulmann S, Sipos B, Kloor M, von Heydebreck A, Schweipert J, et al. Acinar cell carcinomas of the pancreas: a molecular analysis in a series of 57 cases. Virchows Arch. 2014;465:661–72.
Chmielecki J, Hutchinson KE, Frampton GM, Chalmers ZR, Johnson A, Shi C, et al. Comprehensive genomic profiling of pancreatic acinar cell carcinomas identifies recurrent RAF fusions and frequent inactivation of DNA repair genes. Cancer Disco. 2014;4:1398–405.
Wang L, Basturk O, Wang J, Benayed R, Middha S, Zehir A, et al. A FISH assay efficiently screens for BRAF gene rearrangements in pancreatic acinar-type neoplasms. Mod Pathol. 2018;31:132–40.
Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer. 2016;138:881–90.
Chou A, Kim Y, Samra JS, Pajic M, Gill AJ. BRAF gene rearrangements can be identified by FISH studies in pancreatic acinar cell carcinoma. Pathology. 2018;50:345–8.
Chou A, Brown IS, Kumarasinghe MP et al. RET gene rearrangements occur in a subset of pancreatic acinar cell carcinomas. Mod Pathol. 2020;33:657–64.
McEvoy CR, Kee D, Prall OWJ, et al. MEK inhibitor therapy in carcinomas with RAF1 fusions: inferior response in a patient with pancreatic acinar cell carcinoma. JCO Precis Oncol. 2019;3:1–2.
Kim KB, Semrad T, Schrock AB, et al. Significant clinical response to a MEK inhibitor therapy in a patient with metastatic melanoma harboring an RAF1 fusion. JCO Precis Oncol. 2018;2:1–6.
Touat M, Younan N, Euskirchen P, et al. Successful targeting of an ATG7-RAF1 gene fusion in anaplastic pleomorphic xanthoastrocytoma with leptomeningeal dissemination. JCO Precis Oncol. 2019;3:1–7.
McEvoy CR, Xu H, Smith K, et al. Profound MEK inhibitor response in a cutaneous melanoma harboring a GOLGA4-RAF1 fusion. J Clin Invest. 2019;129:1940–5.
McEvoy CR, Semple T, Yellapu B, Choong DY, Xu H, et al. Improved next-generation sequencing pre-capture library yields and sequencing parameters using on-bead PCR. BioTechniques. 2020;68:48–51.
Cameron DL, Schröder J, Penington J, Do H, Molania R, et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017;27:2050–60.
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2.
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92.
Ahdesmäki MJ, Chapman BA, Cingolani P, Hofmann O, Sidoruk A, et al. Prioritisation of structural variant calls in cancer genomes. PeerJ. 2017;5:e3166. https://doi.org/10.7717/peerj.3166. eCollection 2017
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Cutler RE, Stephens RM, Saracino MR, Morrison DK. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci USA. 1998;95:9214–9.
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Picco G, Chen ED, Alonso LG, Behan FM, Gonçalves E, et al. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat Commun. 2019;10:2198.
Hartmaier RJ, Albacker LA, Chmielecki J, Bailey M, He J, et al. High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res. 2017;77:2464–75.
Kato S, Subbiah V, Marchlik E, Elkin SK, Carter JL, Kurzrock R. RET aberrations in diverse cancers: next-generation sequencing of 4,871 patients. Clin Cancer Res. 2017;23:1988–97.
Gao Y, Chang MT, McKay D, Na N, Zhou B, et al. Allele-specific mechanisms of activation of MEK1 mutants determine their properties. Cancer Discov. 2018;8:648–61.
Acknowledgements
The authors gratefully acknowledge Anna Korczynski and Roshana Adeloju (Department of Pathology, Peter MacCallum Cancer Centre), and Peter Diakumis, Sehrish Kanwal, Lavinia Gordon, Jacek Marzec, Tran Pham, and Vladislav Saveliev (University of Melbourne Centre for Cancer Research) for molecular technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
AC is supported by the CINSW ECF (AC0430). This study was supported by the Avner Australian Pancreatic Cancer Genome Initiative (APGI) BioResource partially funded by the Avner Pancreatic Cancer Foundation Grant, www.avnersfoundation.org.au.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Prall, O.W.J., Nastevski, V., Xu, H. et al. RAF1 rearrangements are common in pancreatic acinar cell carcinomas. Mod Pathol 33, 1811–1821 (2020). https://doi.org/10.1038/s41379-020-0545-9
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-020-0545-9
This article is cited by
-
The role of CRAF in cancer progression: from molecular mechanisms to precision therapies
Nature Reviews Cancer (2024)
-
Post-therapy emergence of an NBN reversion mutation in a patient with pancreatic acinar cell carcinoma
npj Precision Oncology (2024)
-
Targeting CRAF kinase in anti-cancer therapy: progress and opportunities
Molecular Cancer (2023)
-
Pancreatoblastomas and mixed and pure acinar cell carcinomas share epigenetic signatures distinct from other neoplasms of the pancreas
Modern Pathology (2022)