Abstract
We report a series of 33 skin tumors harboring a gene fusion of the MAP3K8 gene, which encodes a serine/threonine kinase. The MAP3K8 fusions were identified by RNA sequencing in 28 cases and by break-apart FISH in five cases. Cases in which fusion genes were fully characterized demonstrated a fusion of the 5ʹ part of MAP3K8 comprising exons 1–8 in frame to one of several partner genes at the 3ʹ end. The fusion genes invariably encoded the intact kinase domain of MAP3K8, but not the inhibitory domain at the C-terminus. In 13 (46%) of the sequenced cases, the 3ʹ fusion partner was SVIL. Other recurrent 3ʹ partners were DIP2C and UBL3, with additional fusion partners that occurred only in a single tumor. Clinically, the lesions appeared mainly in young adults (2–59 years of age; median = 18), most commonly involving the lower limbs (55%). Five cases were diagnosed as Spitz nevus, 13 as atypical Spitz tumor, and 15 as malignant Spitz tumor. Atypical and malignant cases more commonly occurred in younger patients. Atypical Spitz tumors and malignant Spitz tumors cases tended to show epidermal ulceration (32%), a dermal component with giant multinucleated cells (32%), and clusters of pigmented cells in the dermis (32%). Moreover, in atypical and malignant cases, a frequent inactivation of CDKN2A (21/26; 77%) was identified either by p16 immunohistochemistry, FISH, or comparative genomic hybridization. Gene expression analysis revealed that MAP3K8 expression levels were significantly elevated compared to a control group of 57 Spitz lesions harboring other known kinase fusions. Clinical follow-up revealed regional nodal involvement in two of six cases, in which sentinel lymph node biopsy was performed but no distant metastatic disease after a median follow-up time of 6 months.
Similar content being viewed by others
Introduction
The modern classification of melanocytic tumors combines clinical, morphological, and genetic features [1]. Among the genetic factors, a unique driver alteration is often identified that initiates clonal expansion and thus is present in all neoplastic cells. These alterations, point mutations or structural rearrangements that result in kinase fusions, commonly lead to constitutive activation of the MAP-kinase pathway and tend to occur in a mutually exclusive pattern with each other. Some mutations are associated with specific histopathological and/or clinical features. The most frequent alterations are mutations that affect residue V600 of BRAF or residues G12, G13, or Q61 of NRAS [2, 3]. These mutations are found in common nevi and are conserved throughout the step by step malignant transformation that can subsequently occur. Tumors related to blue nevi harbor mutations in the Gαq pathway affecting GNAQ, GNA11, PLCB4, or CYSLTR2 [4, 5]. Spitz tumors display a range of alterations with either HRAS mutation or gene fusion involving the kinases ALK, ROS1, NTRK1/3, MET, RET, or BRAF [6,7,8,9,10]. In cutaneous and mucosal melanomas, NF1 and KIT mutations represent other potential driver mutations [11,12,13,14,15]. However, a subset of benign and malignant melanocytic tumors has none of the above mutations and the pathogenically relevant alteration have yet to be identified. A novel fusion involving MAP3K8 was recently described in tumors with a Spitzoid morphology [16, 17]. In this study, we report a series of benign, atypical and malignant cases with MAP3K8 kinase fusions, describe their morphology and identify additional fusion partners.
Materials and methods
Patients and samples
The 33 cases of the cohort were consultation cases from the Department of Biopathology at the Center Léon Bérard in Lyon, France (AF) or cases at University of California, San Francisco in which array comparative genomic hybridization was performed for diagnostic purposes and a copy-number transition within MAP3K8 was identified that made them suspicious for the presence of a fusion. Archival slides stained with hematoxylin and eosin or phloxin and corresponding immunohistochemistry sections were reexamined for notable morphological and immunohistochemical staining features. Morphological analysis and scoring of all cases were performed by AF and IY. Tumor size, growth kinetics, and anatomic location were obtained from the requisition form and array comparative genomic hybridization and MAP3K8 fluorescence in situ hybridization (FISH) results included in the initial report were recorded. For 21 cases clinical follow-up information could be obtained from the referring pathologist or the clinician. The study was conducted according to the Declaration of Helsinki and has been approved by the research ethics committee of the Center Léon Bérard (ref: L18-020) and UCSF (11-07951).
A control group consisted of 57 cutaneous Spitz tumors with other types of oncogenic driver mutations: 17 cases with ALK fusion, 17 cases with ROS1 fusion, 12 cases with NTRK1 fusion, and 11 cases with NTRK3 fusion. Of these, 23 cases were diagnosed as Spitz nevus, 32 as atypical Spitz tumor, and 2 as malignant Spitz tumor.
RNA sequencing
RNA sequencing was performed for 30 cases (for three cases tumor samples were not sufficient). Total RNA was extracted from macrodissected formalin-fixed paraffin-embedded tumor sections using the FormaPure RNA kit (Beckman Coulter #C19158, Brea, CA, USA). RNase-free DNase set (Qiagen #AM2222, Courtaboeuf, France) was used to remove DNA. RNA quantification was assessed using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) measurement and RNA quality using the DV200 value (the proportion of the RNA fragments larger than 200 nt) assessed by a TapeStation with Hs RNA Screen Tape (Agilent, Santa Clara, CA, USA). Samples with sufficient RNA quantity (>0.5 µg) and quality (DV200 > 30%) were further analyzed by RNA sequencing. One-hundred nanograms of total RNA was used to prepare libraries with TruSeq RNA Exome (Illumina #20020183, San Diego, USA). Twelve libraries were pooled at a concentration of 4 nM each together with 1% PhiX. Sequencing was performed (paired end, 2 × 75 cycles) using NextSeq 500/550 High Output V2 kit on a NextSeq 500 machine (Illumina).
The mean number of reads per sample was around 80 million. Alignments were performed using STAR on the GRCh38 version of the human reference genome. Number of duplicate reads were assessed using PICARD tools. Samples with a number of unique reads below 10 million (5 million paired-reads) were discarded from the analysis. Fusion transcripts were called by five different algorithms, including STAR-Fusion, FusionMap, FusionCatcher, TopHat-Fusion, and EricScript. MAP3K8 expression values for were extracted using Kallisto version 0.42.5 tool17 with GENECODE release 23-genome annotation based on GRCh38 genome reference. Kallisto TPM expression values were transformed in log2(TPM+2), and all samples were normalized together using the quantile method from the R limma package within R (version 3.1.1) environment.
Fluorescence in situ hybridization
FISH was performed on 4 μm formalin-fixed paraffin-embedded tissue block sections, using the ZytoLight FISH-Tissue Implementation Kit (Zytovision # Z-2028-20, Bremerhaven, Germany), and the MAP3K8 break-apart probe (Empire Genomics # MAP3K8BA-20ORGR, Amplitech, Compiègne, FR) or the CDKN2A locus-specific probe (Zytovision, # Z-2063-200, Bremerhaven, Germany).
FISH signals were enumerated in at least 50 non-overlapping intact nuclei. For MAP3K8 rearrangement, a specimen was considered positive if >20% of nuclei demonstrated a signal pattern consistent with a gene rearrangement (split of orange and green signals or single orange signals). For CDKN2A deletion, a specimen was considered carrying a homozygous deletion if >50% of nuclei demonstrated a pattern with no green signal.
Immunohistochemistry and special stains
Immunohistochemical staining was performed on 4 µm formalin-fixed paraffin-embedded tissue block sections using a Ventana BenchMark Ultra automated stainer (Ventana, Tucson, USA) and Enhanced Alkaline Phosphatase Red Detection Kit (Ventana #800-031) or UltraView Universal DAB Detection kit (Ventana; #760-500). The following antibodies were used: Anti-melanosome (clone HMB45, 1:100, DAKO), Ki67 (clone SP6, 1:200 and P16 (clone ready-for use, Ventana).
Array comparative genomic hybridization
DNA extraction was performed by macro-dissecting formalin-fixed paraffin-embedded tissue block sections followed by the use of the QIAamp DNA micro kit (Qiagen #56304, Hilden, DE). Fragmentation and labeling were done according to manufacturer's protocol (Agilent Technologies), using 1.5 µg of genomic DNA. Tumor DNA was labeled with Cy5, and a reference DNA (Promega #G1521or #G1471, Madison, USA) was labeled with Cy3. Labeled samples were then purified using KREApure columns (Agilent Technologies #5190-0418). Labeling efficiency was calculated using a Nanodrop ND2000 Spectrophotometer. Co-hybridization was performed on 4x180K Agilent SurePrint G3 Human oligonucleotide arrays (Agilent Technologies #G4449A) or on 4x180K Agilent Sureprint G3 Human oligonucleotide arrays custom (Agilent Technologies #G4125A). Slides were washed, dried and scanned on the Agilent SureScan microarray scanner. Scanned images were processed using Agilent Feature Extraction software V11.5 and the analysis was carried out using the Agilent Genomic Workbench software V7.0.
DNA sequencing
DNA sequencing was performed for six cases in which DNA was available after array comparative genomic hybridization. Sequencing libraries were prepared from extracted DNA using the KAPA HyperPrep Kit per the manufacturer’s instructions (KAPA Biosystems, Wilmington, MA p/n KK8504). Custom-designed bait libraries were used to target the coding regions of 80 melanoma-related genes and the TERT promoter (xGen Lockdown probes, Integrated DNA Technologies, Coralville, Iowa). Sequencing was performed as paired end 100 base pair reads on an Illumina HiSeq 4000 (Illumina, San Diego, CA). Sequencing reads were aligned to the human reference sequence UCSC build hg19 (NCBI build 37), using BWA-MEM 0.7.13 [18]. Variant calling was performed with Freebayes 0.9.20 and Unified Genotyper GATK: Appistry v2015.1.1-3.4.46-0-ga8e1d99, Pindel: 0.2.5b8, and Delly 0.7.2 [19,20,21,22]. Variant annotation was performed with Annovar [23].
Results
In this study, we analyzed the clinical, histopathological, immunohistochemical, and genetic features of 33 cases of melanocytic tumors with MAP3K8 fusion (Tables 1–3, respectively).
Clinical data
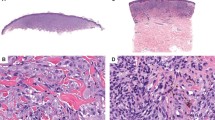
There were slightly more females (n = 18) than males (n = 15) and the age at diagnosis ranged from 2 to 59 years (median 18). The average age of patients with malignant tumors was lower (median 13 years) compared to those with atypical or benign (median 17 and 23 years, respectively). Tumors clinically appeared as exophytic pigmented nodules. Nine cases were ulcerated (32%) (Fig. 1a), of which eight were diagnosed as malignant. Tumors arose at various anatomic sites but were more common on the lower extremities (18/33 or 55%). The clinical diameter of the lesions ranged from 2 to 27 mm (median 6 mm), with atypical and malignant lesions having larger sizes (median of 4.5 mm in Spitz nevus; 6 mm in atypical Spitz tumor; and 7 mm in malignant Spitz tumor). The largest tumor presented as a slowly growing, pigmented nodule on the arm (Fig. 1b and 6) Clinical follow-up information could be obtained for 21 of patients (three Spitz nevi, seven atypical Spitz tumors, and 11 malignant Spitz tumors), with a median follow-up interval of 6 months. Six of the lesions diagnosed as malignant Spitz tumor underwent sentinel lymph node biopsy, of which two were positive. The completion lymphadenectomy did not show additional nodal deposits. No local relapse or widespread metastatic disease was reported.
Melanocytic tumors with MAP3K8 fusions share common morphologic features
Histopathologically, all cases fell into the diagnostic category of Spitz tumors. Five were classified as Spitz nevus, 13 as atypical Spitz tumor, and 15 as malignant Spitz tumor. Representative examples are shown in Figs. 2–4. Most cases had a predominantly nested junctional component (32/33, 97%) and showed hyperplasia of the epidermis (30/33, 90%). Pagetoid scatter was seen in 15 cases (45%) with ascent of large, sometimes multinucleated, epithelioid melanocytes into the upper levels of the epidermis. Ulceration was present in 1/13 atypical Spitz tumors and 8/15 malignant Spitz tumors. Atypical features included cellular nodules in the dermis without maturation. Numerous very large, multinucleated, non-pigmented melanocytes that were scattered diffusely throughout the lesion were present in three atypical Spitz tumors and in six malignant Spitz tumors. The largest multinucleated melanocyte reached close to 200 µm in size (Fig. 4f). Small clusters of heavily pigmented melanocytes surrounded by amelanotic cells in the deep dermis or to the side were present in four atypical Spitz tumors (Fig. 3i, j) and in five malignant Spitz tumors. The clusters were strongly positive by HMB45 immunohistochemistry, against a background of weaker and patchy dermal immunoreactivity (Fig. 5). Desmoplasia was present in 24/33 (73%) cases. A predominantly spindled cytology was mainly found in benign lesions (4/5 Spitz nevi, 1/13 atypical Spitz tumors, and 2/15 malignant Spitz tumor).
Histopathology of Spitz Nevi with MAP3K8 fusion. a, b Symmetrical, well-circumscribed, mainly junctional Spitz nevi. c Close-up view of a with vertically oriented nests of small spindled and pigmented melanocytes with hyperplastic epidermis. d Close-up view of b with vertically oriented nests of large spindled melanocytes below a hyperplastic epidermis and smaller, pigmented nevoid cells at the bottom. e Exophytic Spitz nevus with nests of spindled and epithelioid melanocytes with hyperplastic epidermis. f Close-up view of e. Pagetoid scatter and epithelioid and small clusters of spindled and epithelioid melanocytes in the upper dermis
Atypical Spitz tumors with MAP3K8 fusion. a Dome-shaped, mainly dermal proliferation with a myxoid stroma and lymphocytic infiltrate. b Close-up view of a. Cluster of large multinucleated cells with ample cytoplasm. c Dome-shaped, mainly dermal proliferation with epidermal hyperplasia. d Close-up view of c. Plexiform infiltration of large epithelioid melanocytes without maturation. e Dome-shaped, predominantly dermal proliferation. f Dense dermal nests of large epithelioid melanocytes with giant multinucleated cells. g Regional loss of p16 expression in the more cellular area. h FISH shows loss of CDKN2A (green) compared to reference probe (red). i Dome-shaped, mainly dermal proliferation with deep-dermal nodules. Hyperplasia of the epidermis with central erosion and crust. j Close-up view of deep-nodule with hyperpigmented, spindled, and epithelioid melanocytes
Malignant Spitz tumors with MAP3K8 fusion. a Large exophytic, wedge-shaped proliferation infiltrating deeply into the dermis. b Close-up view of a. Large epithelioid, unpigmented melanocytes with scattered mitotic figures. c Exophytic, mainly dermal proliferation with broad superficial ulceration. Cellular sheets and fascicles of large spindled melanocytes. d–f High-power detail of multiple large multinucleated melanocytes reaching up to 200 microns in size in f
Focal HMB45-positive pigmented cells in Malignant Spitz tumor with MAP3K8 fusion (same case as Fig. 1a). a Exophytic, mainly dermal proliferation with broad superficial ulceration. b HMB45 stain with foci of strong immunoreactivity against a background of patchy dermal positivity. c Close-up view of HMB45-positive area showing a pigmented aggregates of large epithelioid melanocytes. d HMB45 stain of area c
SVIL is the most common fusion partner of MAP3K8
RNAseq identified a chimeric transcript involving MAP3K8 in 28 cases. A MAP3K8-SVIL fusion transcript was identified in 13 cases (46%; Table 1), linking exons 1–8 of MAP3K8 (NM_005204) to either exon 4 (nine cases) or 6 (four cases) of SVIL (NM_003174). MAP3K8 and SVIL reside on chromosome 10 at ~1 megabase distance. A graphical model of the MAP3K8-SVIL fusions is displayed in Fig. 7b. Most malignant tumors with numerous large multinucleated melanocytes (5/7; 71%) or intradermal pigmented nodules (5/7; 71%) had a MAP3K8-SVIL fusion. MAP3K8 was found to be fused to additional 3ʹ partners, two of which were recurrent: DIP2C (three cases) and UBL3 (two cases). In the eight remaining cases, MAP3K8 was fused to STX7, CUBN, SLC4A4, PIP4K2A, CDC42EP3, SFMBT2, CCNY, LINC00703, SPECC1, or MIR3681HG. All fusion transcripts preserved the reading frame of the 3ʹ fusion partner.
Malignant Spitz tumor with MAP3K8 fusion (same case as Fig. 1b). a Dome-shaped, mainly dermal proliferation with deep invasion of the subcutis. b Confluent dermal nests underneath a normal appearing epidermis. The Grenz zone is not obscured. Nests are enveloped by fibrous septa with inflammatory cells. c Close-up view of nests with dense aggregates of spindled and epithelioid, unpigmented melanocytes. A mitotic figure is visible at the left. d Array comparative genomic hybridization profile with losses of several chromosomal regions and gain of chromosome 15
None of the tumors had mutations in BRAF, NRAS, HRAS, or other pathogenic kinase fusions.
MAP3K8 mRNA levels were compared between 30 cases of MAP3K8-fused tumors and 57 Spitz tumors with other kinase fusions such as ALK, ROS1, or NTRK1/3. The expression of MAP3K8 was significantly higher in the MAP3K8-fused group as shown in the box-plot analysis (p = 2.3e−11; Fig. 7c).
MAP3K8 break-apart FISH was positive in 19/24 cases (Fig. 7d), including 4/8 cases with the MAP3K8-SVIL fusion. One tested MAP3K8-UBL3 fusion was also negative. In five cases in which RNA sequencing was not feasible but the morphology or a copy-number transitions of the MAP3K8 locus identified by array comparative genomic hybridization profile suggested a potential MAP3K8 breakpoint, FISH was positive. Among the 19 FISH-positive cases, a rearrangement of MAP3K8 with an unbalanced pattern (one orange/green fusion signal and 1 orange signal per nucleus) was seen in eight cases (37%, including three cases with SVIL partner).
Array comparative genomic hybridization was performed in 12 cases (one Spitz nevus, three atypical Spitz tumors, and eight malignant Spitz tumors) and the frequency of copy-number change is shown in Fig. 7a. The most recurrent copy-number alteration was a loss of chromosome 9, with loss of the short arm found in 75% of cases. Homozygous deletion of CDKN2A at locus 9p21 was identified in four cases (33% of assessed cases). Gains of 10p involving the MAP3K8 locus were the second most frequent copy-number change (66%) and were associated with a copy-number transitions in the MAP3K8 gene. A gain of 7q was found in 50% of cases.
Loss of CDKN2A or its protein product p16 were assessed by FISH, array comparative genomic hybridization, or immunohistochemistry, in a total of 29 cases. Alterations were absent in the three Spitz nevi, but present in 10/13 atypical Spitz tumors and 11/13 malignant Spitz tumors. Homozygous deletion of CDKN2A by array comparative genomic hybridization or FISH was more common in malignant Spitz tumors (7/8 cases) compared to atypical Spitz tumors (2/10 cases).
The TERT promoter was genotyped in six cases (one Spitz nevus, three atypical Spitz tumors, two malignant Spitz tumors). A single TERT promoter hotspot mutation was identified (g.1295250C > T) in a malignant Spitz tumor.
Discussion
MAP3K8 is a serine threonine kinase that was described by Miyoshi et al. [24] as an oncogene isolated from a thyroid cancer cell line. They named this gene COT for Cancer Osaka Thyroid and proved it had an in vitro transforming capacity when artificially introduced into the SHOK hamster embryonic cell line. In parallel, studies identified the gene as the target of a viral insertion in the 3ʹ end that results in a fusion protein with an altered C-terminal end, which leads to T-cell transformation in rats [25]. This gene also had an in vitro transforming capacity of the NIH 3T3 fibroblasts [24]. Similarly, another viral insertion that resulted in a C-terminal-truncated protein was found to induce mammary carcinomas in mice [26]. different mechanisms have been found to explain the increased expression of the fusion transcript and the transforming ability of the C-terminally truncated gene. Firstly, the kinase activity of the protein is increased, related to the removal of a potentially folded C-terminal portion covering the kinase domain [27, 28]. Secondly, C-terminal truncation removes a proteasomal proteolytic site (located in aa positions 435–457) [29]. Hypothetically, regulatory microRNAs failing to bind to the missing C-terminal portion of MAP3K8 mRNA could also be another mechanism, leading to increased protein expression [30]. Moreover, MAP3K8 overexpression has also more recently been described as an oncogenic event found in ovarian and squamous cell carcinomas [31, 32]. Also, melanomas treated with BRAF inhibitors can develop secondary resistance due to MAP3K8 overexpression, which results in MEK activation, independent of BRAF [33].
Two recent studies have described genomic alterations involving MAP3K8 in a subset of cutaneous melanocytic tumors [16, 17]. A total of 17 different fusions and five truncating mutations in MAP3K8 have been identified, all events compromising its C-terminal domain. If we add the results from our 28-sequenced cases, the most frequent fusion partner is the supervillin gene (SVIL) representing nearly half of the cases (20/45 = 44%). SVIL is a very large actin-binding protein with 5 Gelsolin homology domains and a high capacity to bind to the plasma membrane and the actin cytoskeleton [34]. In our series, 9 of the SVIL-fused cases had fusions in which the SVIL portion started with exon 4 and four cases in which it started with exon 6. Since the 3ʹUTR of SVIL spans from exon 1 to 6, it is likely that the resulting chimeric protein includes the entirety of SVIL, likely resulting in a still functional supervillin protein component. Supervillin promotes proliferation and migration in various cell lines and, therefor,e could contribute to the oncogenic effects of the fusion protein [35]. Other 3ʹ fusion partners identified included: DIP2C (6/45), UBL3 (4/45), STX7 (2/45), SPECC1 (2/45), CUBN (2/45). Other single 3ʹ fusion partners were identified either in this study (SFMBT2, MIR3681HG, CDC42EP, SLC4A4, CCNY, LINC00703, and PIP4K2A are seven novel 3ʹ partners) or in the previous studies (GNG2, PRKACB and PCDH7). No clear homology in terms of function, binding capacity, or protein size were identified in these 3ʹ partners.
Newman et al. [17] have confirmed the transforming ability of MAP3K8-fused or truncated in melanocytes. Our analysis shows that the fusion is associated with increased expression levels compared to Spitz tumors with other fusions. This could reflect increased stability of the transcript, gene dosage effects as we observed copy-number increases in a subset of cases, or yet to be identified changes in MAP3K8 gene regulation.
Our study also focused on clinical and morphological features. Several clinical elements stood out. Firstly, an early onset of malignant transformation could occur, possibly driven by CDKN2A. This is a known secondary event in melanoma progression, that has been well established both in the group of Spitz tumors or in melanomas arising from common nevi [36, 37]. Secondly, the preferred anatomic location was the leg. All six cases from the study of Quan et al. were on the lower limbs, as well as the malignant Spitz tumor case described by Newman et al. While Spitz nevi in general are common in this location [38], the anatomic site distribution as a function of oncogenic driver mutation has not been studied. Thirdly, superficial ulceration was seen in more than half of the malignant Spitz tumors and mainly in MAP3K8-SVIL tumors (5/7). This feature is rather uncommon in malignant Spitz tumor, and often attributed to trauma on the limbs of children. Similarly, both of the MAP3K8-SVIL fused case reported by Quan et al, were ulcerated, on the limbs and one was a young child [16]. In previous reports of genetically defined Spitz tumors with ALK, ROS1, NTRK1, and NTRK3 fusions, ulcerations were rarely described (16% at most of ALK-fused atypical Spitz tumors and malignant Spitz tumors) compared to a 71% ulceration frequency in the MAP3K8-SVIL subgroup [39]. These clinical and histopathological features could help predict the genetic driver, if confirmed in wider studies.
Several morphological features were noticeable in our study. Firstly, the cytology, was predominantly spindled in the junctional nests of Spitz nevi but was mostly epithelioid in the dermal component of atypical Spitz tumors and malignant Spitz tumors. Moreover, the presence of diffuse giant multinucleated melanocytes was detected in the atypical Spitz tumor group (3/12) and became more frequent in the malignant Spitz tumor group (6/14). These large, sometimes giant melanocytes reaching up to 200 µm in size have not so far been clearly described or linked to a specific subtype of Spitz nevus. They have not been reported with other genetic anomalies and had resonance with the original description of Spitz nevus. In her 1948 paper, Melanomas of Childhood, Sophie Spitz stated that “In one feature alone some of these lesions were distinctly different from the malignant melanoma of adults. In eight of the nine cases just described, giant cells were present both in the epidermal and dermal portion of the tumor (Fig. 3 and 4). In five cases there were small to moderate numbers of these cells, but in three cases giant cells were present in such large numbers as to constitute the most outstanding feature of the lesion” [40]. It seems likely, given the percentage of borderline Spitzoid neoplasms with MAP3K8 fusions that some of these cases were examples of this entity. Quan et al. underscored the predominance of the epithelioid subtype in the dermis in their six cases, but did not specifically mention giant multinucleated cells. Secondly, focal hyperpigmented dermal clones were seen in atypical Spitz tumors (4/12), and malignant Spitz tumors (5/14). They were not cytologically different from the surrounding melanocytes, besides their pigmentation. Stronger HMB45 staining often highlighted these areas. Again, they tended to be more frequent in the MAP3K8-SVIL subgroup (5/13). They were also noted by Quan et al. in 3/6 cases (including one MAP3K8-SVIL). Further, morphological comparative studies between groups of Spitz tumors bearing the various types of gene fusions could establish if these morphological features are specific of MAP3K8.
Molecular findings. a Frequency plot chromosomal aberrations as determined by array comparative genomic hybridization. Gains are shown in red and losses in green. b Graphical view of two MAP3K8-SVIL fusions involving with a breakpoint either before exon 4 or 6 of SVIL. c Box-plot expression levels of MAP3K8 from RNAseq data in MAP3K8-fused and tyrosine kinase-fused Spitz tumors. d MAP3K8 FISH break-apart probe showing disruption of one of the signal pairs (arrows) with loss of one green signal Insert showing mapping of FISH probes (Case no. 10 with MAP3K8-SVIL fusion)
MAP3K8 and SVIL are separated by about 1Mb and oriented in opposite directions within the short arm of chromosome 10. The MAP3K8-SVIL fusion gene could, therefore, arise through a paracentric inversion of this chromosomal region. According to this hypothesis, the rearrangement of MAP3K8 in this specific fusion transcript could interfere with detection by FISH using a break-apart probe strategy. MAP3K8 rearrangements were readily detectable in half of the eight MAP3K8-SVIL tested cases, with either a large separation of green and orange signals or an unbalanced pattern. The unbalanced pattern was not associated with a specific partner gene or any other clinical or morphological characteristic of the cohort. Overall, the FISH technique was able to detect 19/24 (79%) cases of MAP3K8-fused tumors and can therefore be used as a cost-effective way of screening for MAP3K8 rearrangements in routine as there is currently no reliable immunohistochemistry test to detect the anomaly.
Quan et al. [16] reported nodal involvement in one case and a local recurrence in another case. Newman et al. [17] reported a case with extensive involvement of a limb, in which MEK-targeting therapy transiently controlled the local disease, but failed to prevent progression metastatic dissemination to lungs and bone with subsequent death. Although no widespread metastatic disease was observed in our cases, a longer follow-up is needed to more clearly evaluate the prognosis of this new molecular subtype. In our series, malignant cases were predominant (45%) over atypical (40%) and benign lesions (15%). While our assessment of TERT promoter status was limited to six cases, we identified a hotspot TERT promoter mutation in one of two malignant lesions, but not in three atypical and one benign lesions. The lethal case reported by Newman et al. also demonstrated dysregulation of TERT (by complex structural rearrangement). These findings suggest that TERT dysregulation could be associated with malignant behavior.
Moreover, since a TCGA analysis identified potential oncogenic MAP3K8 alterations in 1.5% of samples and that truncating mutations were also identified in uterine endometrioid carcinoma and colorectal adenocarcinomas with a hotspot found at position R442, we explored the presence of this alteration in other types of malignant tumors we had studied in our institution by RNA sequencing. We identified fusions of MAP3K8 exon 8, removing the C-terminal portion of the protein, in the metastasis of a high-grade serous adenocarcinoma of the ovary, as well as in two mesotheliomas and a cutaneous myxo-inflammatory fibroblastic sarcoma (data not shown), emphasizing that this oncogenic alteration is not restricted to melanocytic neoplasms.
Altogether, we report an extended series of melanocytic tumors harboring a serine/threonine kinase fusion involving MAP3K8, presenting as Spitz tumors with a spectrum ranging from benign to malignant. Our findings indicate that the range of possible 3ʹ fusion partners is wide but that MAP3K8-SVIL fusions are most common. Histopathological features that can alert the pathologist to the presence of a MAK3K8 fusion is the presence of large multinucleated cells in the dermis and ulceration.
References
Elder DE, Massi D, Scolyer R, Willemze R. WHO classification of skin tumours. [cited 4 November 2018]. Available from: http://publications.iarc.fr/Book-And-Report-Series/Who-Iarc-Classification-Of-Tumours/WHO-Classification-Of-Skin-Tumours-2018.
Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127:179–82.
Papp T, Schipper H, Kumar K, Schiffmann D, Zimmermann R. Mutational analysis of the BRAF gene in human congenital and dysplastic melanocytic naevi. Melanoma Res. 2005;15:401–7.
Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602.
Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9.
Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000;157:967–72.
Botton T, Yeh I, Nelson T, Vemula SS, Sparatta A, Garrido MC, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 2013;26:845–51.
Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116.
Yeh I, Botton T, Talevich E, Shain AH, Sparatta AJ, de la Fouchardiere A, et al. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun. 2015;6:7174.
Yeh I, Tee MK, Botton T, Shain AH, Sparatta AJ, Gagnon A, et al. NTRK3 kinase fusions in Spitz tumours. J Pathol. 2016;240:282–90.
Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–6.
Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002.
Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96.
Ablain J, Xu M, Rothschild H, Jordan RC, Mito JK, Daniels BH, et al. Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science. 2018;362:1055–60.
Yeh I, Jorgenson E, Shen L, Xu M, North JP, Shain AH, et al. Targeted genomic profiling of acral melanoma. J Natl Cancer Inst. 2019. https://doi.org/10.1093/jnci/djz005.
Quan VL, Zhang B, Mohan LS, Shi K, Isales MC, Panah E, et al. Activating structural alterations in MAPK genes are distinct genetic drivers in a unique subgroup of spitzoid neoplasms. Am J Surg Pathol. 2019;43:538–48.
Newman S, Fan L, Pribnow A, Silkov A, Rice SV, Lee S, et al. Clinical genome sequencing uncovers potentially targetable truncations and fusions of MAP3K8 in spitzoid and other melanomas. Nat Med. 2019. https://doi.org/10.1038/s41591-019-0373-y.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–9.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–33.
Garrison EMG. Haplotype-based variant detection from short-read sequencing. arXiv. 2012;1207:3907
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl Acids Res. 2010;38:e164–4.
Miyoshi J, Higashi T, Mukai H, Ohuchi T, Kakunaga T. Structure and transforming potential of the human cot oncogene encoding a putative protein kinase. Mol Cell Biol. 1991;11:4088–96.
Makris A, Patriotis C, Bear SE, Tsichlis PN. Genomic organization and expression of Tpl-2 in normal cells and Moloney murine leukemia virus-induced rat T-cell lymphomas: activation by provirus insertion. J Virol. 1993;67:4283–9.
Erny KM, Peli J, Lambert JF, Muller V, Diggelmann H. Involvement of the Tpl-2/cot oncogene in MMTV tumorigenesis. Oncogene. 1996;13:2015–20.
Ceci JD, Patriotis CP, Tsatsanis C, Makris AM, Kovatch R, Swing DA, et al. Tpl-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Genes Dev. 1997;11:688–700.
Gándara ML, López P, Hernando R, Castaño JG, Alemany S. The COOH-terminal domain of wild-type Cot regulates its stability and kinase specific activity. Mol Cell Biol. 2003;23:7377–90.
Gantke T, Sriskantharajah S, Ley SC. Regulation and function of TPL-2, an IκB kinase-regulated MAP kinase kinase kinase. Cell Res. 2011;21:131–45.
Zhang X, Jiang P, Shuai L, Chen K, Li Z, Zhang Y, et al. miR-589-5p inhibits MAP3K8 and suppresses CD90+ cancer stem cells in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:176.
Gruosso T, Garnier C, Abelanet S, Kieffer Y, Lemesre V, Bellanger D, et al. MAP3K8/TPL-2/COT is a potential predictive marker for MEK inhibitor treatment in high-grade serous ovarian carcinomas. Nat Commun. 2015;6:8583.
Lee J-H, Lee J-H, Lee SH, Do S-I, Cho S-D, Forslund O, et al. TPL2 is an oncogenic driver in keratocanthoma and squamous cell carcinoma. Cancer Res. 2016;76:6712–22.
Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72.
Pope RK, Pestonjamasp KN, Smith KP, Wulfkuhle JD, Strassel CP, Lawrence JB, et al. Cloning, characterization, and chromosomal localization of human superillin (SVIL). Genomics. 1998;52:342–51.
Chen X, Yang H, Zhang S, Wang Z, Ye F, Liang C, et al. A novel splice variant of supervillin, SV5, promotes carcinoma cell proliferation and cell migration. Biochem Biophys Res Commun. 2017;482:43–9.
Yazdan P, Cooper C, Sholl LM, Busam K, Rademaker A, Weitner BB, et al. Comparative analysis of atypical spitz tumors with heterozygous versus homozygous 9p21 deletions for clinical outcomes, histomorphology, BRAF mutation, and p16 expression. Am J Surg Pathol. 2014;38:638–45.
Shain AH, Bastian BC. The genetic evolution of melanoma. N Engl J Med. 2016;374:995–6.
Lazova R, Pornputtapong N, Halaban R, Bosenberg M, Bai Y, Chai H, et al. Spitz nevi and Spitzoid melanomas—exome sequencing and comparison to conventional melanocytic nevi and melanomas. Mod Pathol. 2017;30:640–9.
Yeh I, de la Fouchardiere A, Pissaloux D, Mully TW, Garrido MC, Vemula SS, et al. Clinical, histopathologic, and genomic features of spitz tumors with ALK fusions. Am J Surgical Pathol. 2015;39:581–91.
Spitz S. Melanomas of childhood. Am J Pathol. 1948;24:591–609.
Acknowledgements
We thank the following pathologists for their case contribution: Drs. Paoletti, Plantier, de Mauroy, Niel, Habougit, Michiels marzais, Ziadé, Castillo, Petit, Vacheret, Voulat, Ducoin, Macagno, Palau, Rethers, Doumes-Lacoste, Gosset, Neumann, Palasse, Laurent-Roussel, Terrier, Arcin-Thoury, Carr, Godeneche, Martin de la Salle, and the following physicians for clinical and follow-up data: Drs. de Quatrebarbe, Diner, Chedeville, Pecheux, Perrot, Sage, Hauguel, Meyer, Rossi-Chouquet, Poeuf, Gillibert, Meyer, Berbis, Parrocel, Armingaud, Irani, Poddevin, Levy-Roi, de Berail, Chamain, Safia, Monteanu, Duval, Mortier, Dalle. Boris Bastian is supported by a grant from the National Cancer Institute (1R35CA220481). We also thank Florine Dreux, Elise Malandin, Cyril Py, Marlène Perrin-Niquet, Colline Taillandier, and Elodie Legrand for their technical help. We thank Sandrine Paindavoine and Pr Françoise Galateau for their assistance with the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Houlier, A., Pissaloux, D., Masse, I. et al. Melanocytic tumors with MAP3K8 fusions: report of 33 cases with morphological-genetic correlations. Mod Pathol 33, 846–857 (2020). https://doi.org/10.1038/s41379-019-0384-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0384-8
This article is cited by
-
Novel gene fusion discovery in Spitz tumours and its relevance in diagnostics
Virchows Archiv (2023)
-
Novel insights into the BAP1-inactivated melanocytic tumor
Modern Pathology (2022)
-
Morphologic features in a series of 352 Spitz melanocytic proliferations help predict their oncogenic drivers
Virchows Archiv (2022)
-
Clinical, morphologic, and genomic findings in ROS1 fusion Spitz neoplasms
Modern Pathology (2021)
-
GOPC-ROS1 mosaicism in agminated Spitz naevi: report of two cases
Virchows Archiv (2021)