Abstract
The switch from in situ to invasive tumor growth represents a crucial stage in the evolution of lung adenocarcinoma. However, the biological understanding of this shift is limited, and ‘Noguchi Type C’ tumors, being early lung adenocarcinomas with mixed in situ and invasive growth, represent those that are highly valuable in advancing our understanding of this process. All Noguchi Type C adenocarcinomas (n = 110) from the LATTICE-A cohort were reviewed and two patterns of in situ tumor growth were identified: those deemed likely to represent a true shift from precursor in situ to invasive disease (‘Noguchi C1’) and those in which the lepidic component appeared to represent outgrowth of the invasive tumor along existing airspaces (‘Noguchi C2’). Overall Ki67 fraction was greater in C2 tumors and only C1 tumors showed significant increasing Ki67 from in situ to invasive disease. P53 positivity was acquired from in situ to invasive disease in C1 tumors but both components were positive in C2 tumors. Likewise, vimentin expression was increased from in situ to invasive tumor in C1 tumors only. Targeted next generation sequencing of 18 C1 tumors identified four mutations private to the invasive regions, including two in TP53, while 6 C2 tumors showed no private mutations. In the full LATTICe-A cohort, Ki67 fraction classified as either less than or greater than 10% within the in situ component of lung adenocarcinoma was identified as a strong predictor of patient outcome. This supports the proposition that tumors of all stages that have ‘high grade’ in situ components represent those with aggressive lepidic growth of the invasive clone. Overall these data support that the combined growth of Noguchi C tumors can represent two differing biological states and that ‘Noguchi C1’ tumors represent the genuine biological shift from in situ to invasive disease.
Similar content being viewed by others
Introduction
Invasive carcinomas of the lung arise from in situ precursors [1]. When precursor cells invade across the basement membrane, they gain access to interstitial tissue and to vascular structures, enabling distant metastasis. This switch from in situ to invasive growth is crucial in the development of lethal tumors, and presents a tempting target for prevention or intervention, especially in high-risk groups.
In comparison to other common adenocarcinomas, preinvasive lung lesions have proved to be difficult to characterize. They can only be accessed following invasive surgery, and the majority of resected tumors are very advanced, in biological terms, and already show widespread invasive behavior. Nonetheless, there is a well-described histopathological continuum of lesions from atypical adenomatous hyperplasia, through adenocarcinoma in situ, then minimally invasive adenocarcinoma, and finally to extensively invasive disease, and this has been adopted into the pathological criteria for diagnosis defined by the WHO and IASLC [2]. This morphological classification is highly prognostic, and tumors with no evidence of invasion have very low rates of nodal metastasis, and very good outcomes postsurgery [3]. However, in comparison to other common malignancies, this ‘Vogelgram’ is not yet accompanied by a detailed catalog of driving genomic changes. Our current knowledge is predominantly based on studies that compare invasive and in situ tumors [4,5,6] that show EGFR amplification copy number gain, TP53 mutation and increasing chromosomal instability identified as genomic changes associated with invasive tumors. More recent large-scale studies have generated illuminating data on tumor evolution by the multiregional sequencing of invasive tumors and metastases [7, 8]. Few studies however have as yet shown evidence of molecular progression from histologically defined in situ to invasive growth within individual tumors. The limited published data shows EGFR amplification and increasing chromosomal rearrangements to be the most conclusively defined changes seen in the transition from in situ to invasive disease [9, 10]. This kind of evidence is vital, as it directly links genomic events to cell biology in single tumors, giving concrete examples of the diverse routes to invasiveness taken by single tumor clones under the influence of natural selection.
We therefore set out to use a rigorous microscopic histopathological approach to identify mutations which are found only in invasive parts of very early lung adenocarcinomas, simultaneously aiming to improve our understanding of tumor morphology in these cancers.
This approach required the identification of early adenocarcinomas containing both areas of in situ precursor and more advanced invasive disease. In living patients this stage is likely to be transient; even small tumors can be wholly invasive, as the aggressive, biologically advanced invasive component often quickly overwhelms the in situ precursor. For this reason, it is necessary to examine large numbers of archival tumors to identify those which show both precursor and invasive modes of growth.
The study performed by Noguchi et al. in 1995 is perhaps the most influential investigation of morphological progression in lung cancer [11]. In this landmark paper, it was shown that clinically early (<20 mm) tumors could be meaningfully separated into six classes designated A–F: A are wholly in situ, B are biologically in situ with areas of stromal alteration morphologically mimicking invasion (‘pseudoinvasion’), C are of mixed in situ and invasive patterns, and D–F are wholly invasive. The four classes are impressively diverse in their survival outcomes, and a descendent of this morphological approach has been incorporated into the current WHO tumor classification and cancer staging. “Noguchi C” lesions present us with an opportunity to sequence in situ and invasive areas from the same tumor, with the hope of identifying associated DNA changes.
However, we believed it probable that not all type C lesions were suitable for the detection of biological progression. It is known that invasion-competent cells can sometimes grow in a lepidic manner, as is sometimes seen in invasive tumors which have metastasized to the lung from other primary organs [12, 13]. This therefore raises the possibility that primary lung adenocarcinoma cells with invasive potential may also opportunistically grow lepidically, giving an in situ appearance. In the same way that Noguchi et al. morphologically separate true invasion from pseudoinvasion by separating class B from class C, we suggest that it is possible to morphologically separate true precursor in situ growth from biologically advanced in situ growth by criteria which are detailed below. Retaining the Noguchi letter classification, we name these two classes C1 and C2: Noguchi C1 tumors are truly biologically biphasic, with a ‘precursor’ in situ component and the potential to reveal genomic changes which drive invasion, while in Noguchi C2 tumors, the in situ component represents opportunistic in situ/lepidic outgrowth by an invasion-competent clone of cells. This carries important clinical implications, as the recognition of in situ growth patterns with very different invasion potential may alter the interpretation of biopsies containing only in situ tumor, or of high-resolution CT imaging showing ground glass changes suggestive of in situ tumor growth.
In this way we identified numerous C1 and C2 lesions from our historical cohort of 994 primary lung adenocarcinomas. We constructed focussed tissue microarrays of in situ and invasive areas from these tumors for quantitative in situ assays of proliferation and p53 expression. Furthermore, in order to detect genomic changes related to step changes in tumor morphology, we performed laser capture microdissection (LCM) of neighboring epithelial areas followed by DNA extraction and next-generation sequencing of a panel of cancer driving genes.
Methods
For this study we interrogated a 994-patient retrospective cohort of resected lung adenocarcinomas for which comprehensive clinicopathological data including survival information has been collated (LATTICe-A: Leicester Archival Thoracic Tumor Investigatory Cohort – Adenocarcinoma). This study was approved by the Northampton Research Ethics Committee (reference 14/EM/1159) and University Hospitals Leicester NHS Trust Research and Innovation Department (reference UHL 11363). The cohort includes all resected primary adenocarcinomas from our surgical center from 1998 to 2014. Nodal invasion status was collected from histopathology reports. Patient outcomes were obtained from local and national databases.
Morphological classification
Two hundred and forty-two tumors ≤20 mm were examined. Scanned digital images (Hamamatsu Nanozoomer™) of all original diagnostic slides were reviewed by two subspecialist thoracic histopathologists (DM and JLQ) and classified as per Noguchi et al. [11]. The pathologists were blinded to the clinical outcome data.
One hundred and ten Noguchi C tumors were identified, defined as tumors with areas of lepidic growth and ‘true’ invasion with fibroblastic stromal changes. Noguchi C tumors were subclassified into class C1 (precursor) and C2 (outgrowth) patterns on morphological grounds: we applied a simple score to each tumor, giving one point each for observation of: (i) clear nuclear grade difference (size/shape/pleomorphism) between the in situ and invasive components, (ii) architectural asymmetry reflecting the centrality of the invasive component and (iii) the absence of an in situ ‘penumbra’, with penumbra referring an in situ component of uniform width at the edge of the lesion (Fig. 1a). Tumors scoring 3 or 2 were classified as ‘Noguchi C1’, while those scoring 1 or 0 were classified as ‘Noguchi C2’. Examples of each type are shown in Fig. 1b.
a Schematic representation of the morphological characteristics of C1 versus C2 tumors. C1 tumors show sporadic appearance of a high-grade invasive focus from a low-grade precursor and are often asymmetrical. As such the typical C1 is lepidic predominant with a minor invasive component, although tumors with any residual lower grade, nonsymmetrical lepidic component would similarly be classified as C1 by our scoring. C2 tumors represent high-grade invasive carcinomas with peripheral lepidic spread (outgrowth) of invasive tumor cells. As C2 tumors are expanding from a central invasive region, they show more symmetry, often with a marked constant-thickness ‘penumbra’ of in situ disease due to lepidic outgrowth. b Histology of representative C1 and C2 tumors. Low-power views show tumor architecture alongside high power regions from the in situ and invasive component of both tumors. c Lymph node positivity at resection by subtype. C2 tumors are more likely to show nodal metastasis than C1 tumors (p < 0.01). c Kaplan–Meier plot of 5 year overall survival of early lung adenocarcinoma patients in the LATTICe-A cohort – Noguchi A and B (biologically in situ), Noguchi C1, Noguchi C2, and a combined group of fully invasive subtypes (d)
Tissue microarray construction
Two tissue microarray series were constructed, the first was a focused Noguchi C microarray containing 44 tumors, the second was an entire lung adenocarcinoma cohort microarray containing 966 tumors.
The Noguchi C tissue microarray was constructed from all Noguchi C tumors with sufficient tissue available. 27 C1 and 17 C2 tumors were included. For each tumor, 2 × 1 mm cores of invasive tumor and 2 × 1 mm cores of in situ tumor were taken.
The full LATTICe-A cohort tissue microarray consisted of 966 tumors from a 994 tumor cohort. 3 × 1 mm cores were obtained from each case. Where possible, cores were taken from different donor blocks, and were selected to maximize sampling of observed variance in tumor pattern. In particular, where possible, both in situ and invasive regions were sampled. Core sampling was directed by examination of scanned whole section H&E images and correspondence to paraffin blocks. Recipient blocks contain either 60 or 40 cases (i.e. 180 or 120 tumor cores) plus orientation and control cores. The entire cohort is represented in 22 tissue microarrays. All tissue microarrays were constructed in quadruplicate using a semiautomated TMArrayer™ (Pathology Devices). Sections were stained with H&E, and all cores were assigned a growth pattern as per IASLC recommendations [2].
Immunohistochemistry
Immunohistochemistry for Ki67, p53, and vimentin was applied to the Noguchi C tissue microarray. This was performed following use of the Dako Target Retrieval Solution pH9, S2375, in a PTLink at 97° for 20 min. The antibodies Ki-67 IR622, p53 IR616, and Vim IR630 were applied and visualized using Dako Flex+ kit K8002, on a Dako Link48 Autostainer. Ki67 staining was also applied to the full LATTICE cohort tissue microarray using the same method.
Manual scoring was performed for all three stains. Ki67 was scored as a percent of nuclear positivity within tumor cells. Scoring for nuclear tumor cell p53 staining was performed as previously described for the reporting of mutant status in lung adenocarcinoma [14]. Vimentin expression in tumor cell was quantified using H-score method (0–300). Individual cores were scored and the mean was taken across cores from a single tumor to determine the whole tumor values.
ALK and ROS1 rearrangement testing was applied to whole sections from the Noguchi C next generation sequencing series only. The Ventana ALK (D5F3) immunohistochemistry assay and Cell Signaling Technology ROS1 (D4D6) assay were used. Both were applied on the Ventana Ultra platform within a clinically accredited molecular pathology laboratory.
Laser capture microdissection
Eighteen C1 tumors and 5 C2 tumors with sufficient residual tissue in the resection blocks following tissue microarray construction underwent LCM using the Zeiss PALM system to separately sample the in situ and invasive components. Serial 10 micron sections were cut from the formalin fixed paraffin embedded block, dewaxed and stained with cresyl violet. Four to ten sections were used for microdissection. DNA was extracted using the Qiagen Generead FFPE DNA extraction kit, producing DNA quantities following extraction of 20–5000 ng. Germline DNA was extracted from uninvolved lymph node tissue.
Next generation sequencing
All samples generated underwent library preparation and targeted next generation sequencing using the Life Technologies Lung and Colon Cancer Panel on an IonTorrent Personal Genome Machine using 10 ng of extracted DNA. This gene panel covers commonly mutated regions of 22 lung and colon cancer associated genes. Driving mutations were identified using the Life Technology Variant Caller software and were identified as either being shared between the invasive and in situ components or private to one of the regions. Full details of the amplicons covered by this gene panel are given in Supplementary Data File 1.
Statistical methods
Following the use of Shapiro–Wilk test of normality for Ki67 and vimentin scores, comparisons between C1 and C2 tumors were performed using the Mann–Whitney test, and between in situ and invasive components of the same tumor using the Wilcoxon signed-rank test. Due to the binary nature of the p53 scores, comparisons between C1 and C2 tumors were performed using the Pearson chi-squared test, and between in situ and invasive components of the same tumor using McNemar’s test. Survival analysis was performed using Kaplan–Meier methods, and significance of difference between groups was assessed by the log-rank test. All survival analysis used an Overall Survival model, which records all patient deaths as events and censors other patients at their last visit date. All statistical analysis was performed using Stata/SE 15.1.
Results
Morphological Noguchi C tumor assessment
Two hundred and forty-two adenocarcinomas from the LATTICe-A cohort (n = 994) were 20 mm or less in size. On pathological review 110 showed both invasive and in situ disease and were therefore classified as Noguchi type C tumors.
Inspection of the Noguchi C tumors revealed two distinct classes based on the appearance of the in situ component. One class of lesions had little symmetry and often showed a higher nuclear grade in the invasive area. We hypothesized that these tumors were showing true biological progression from a low-grade precursor clone to an invasive subclone, and designated them ‘C1’. The other class showed more symmetry, with a constant-thickness peripheral zone (or ‘penumbra’) of lepidic growth surrounding a zone of invasion, with both the invasive and lepidic cells showing similar nuclear features. We hypothesized that in these tumors, the lepidic zone represented opportunistic lepidic peripheral outgrowth of an invasion-competent tumor clone, and designated them ‘C2’. These appearances are summarized in Fig. 1a, b. This detailed classification scheme was applied to all 110 Noguchi C tumors, identifying 42 C1 and 68 C2 tumors. The result of this scoring is presented in Table 1. Demographic and pathological information for these patients is presented in Table 2 and breakdown of tumors types by current WHO classification is presented in Table 3. Just over half of the C1 tumors (22/42) were lepidic predominant (lepidic predominant adenocarcinoma plus minimally invasive adenocarcinoma), whereas only 8 of the 68 C2 tumors fell into either of these categories, with the majority representing acinar predominant adenocarcinoma.
According to our model, C1 tumors were resected when they had only just acquired focal invasive capability and would therefore have had limited opportunity for metastasis. C2 tumors, however, represent established invasive disease. In support of this, the rate of nodal metastasis at resection was significantly higher in C2 tumors (26%) than in C1 tumors (8%) (p = 0.03) (Fig. 1c), although we did not detect a significant difference in patient outcome (Fig. 1d).
Molecular phenotypes of C1 and C2 tumors
Within the focused Noguchi C tissue microarray quantitative in situ immunohistochemical assays were applied. First, we assessed proliferation rates in in situ and invasive parts of both C1 and C2 tumors by Ki67 staining (Fig. 2a). C2 tumors show significantly greater proliferation rates than C1 tumors overall (mean C1 Ki67 8.9% [±1.6] versus C2 23.3% [±2.8], p < 0.0001). Furthermore, the invasive areas of C1 tumors are significantly more proliferative than neighboring in situ areas (10.8% [±1.7] versus 7.1% [1.6], p < 0.001), while invasive and in situ areas of C2 tumors are not significantly different (24.5% [±3.0] versus 22.1% [±2.8], p = 0.3).
a Distribution of Ki67 index scoring on the C1 and C2 tissue microarray. C2 tumors show significantly higher proliferation index than C1 tumors. Also, C1 tumors show a significant increase in proliferation from in situ to invasive disease which is not seen in C2 tumors. b Likewise positivity for p53 staining was significantly higher in C2 lesions overall and showed an increase from in situ to invasive disease only within C1 lesions. c Studying mesenchymal Vimentin as a marker of epithelial–mesenchymal transition within this cohort, expression increased significantly from in situ to invasive disease in C1 tumors only
The difference in proliferation rate between in situ and invasive regions of C1 tumors supports the hypothesis that it is these tumors which are most likely to truly represent biological progression. The much higher proliferation rate of in situ growth in C2 tumors compared to the in situ growth in C1 strongly supports our hypothesis that there are distinct modes of in situ tumor cell proliferation.
TP53 mutations have previously been linked to biological progression in lung adenocarcinoma [14]. Therefore, if our classification of C1 tumors is truly identifying cases undergoing step progression, then we might expect to find a higher rate of mutant p53 in invasive areas of this group. This was assessed in situ by immunohistochemical staining to detect the nuclear accumulation of mutant protein.
C1 tumors showed a lower rate of p53 mutant phenotype positivity than C2 tumors overall, although this was not statistically significant. C1 tumors did however show the acquisition of p53 positivity in 5 of the 27 tumors assessed representing a significant difference between the in situ and invasive (P < 0.05), while all C2 tumors positive in the invasive component were also positive in the in situ component (Fig. 2b).
Finally, we assessed the expression of vimentin as a marker of transition, which is associated with the switch to invasiveness in many solid tumors [15, 16]. Invasive tumor areas showed greater vimentin expression compared to adjacent in situ regions in C1 tumors (mean H-score 88.8 [±23.1] versus 31.4 [±13.8], p < 0.01), with no significant difference in C2 tumors (Fig. 2c).
Representative tissue microarray core images from the staining of all three antibodies are shown in Fig. 3.
Representative staining of in situ and invasive 1 mm diameter cores from the early adenocarcinoma LATTICe-A tissue microarray, for Ki67, p53, and Vimentin. These example cases show Ki67 which is of higher fraction in the invasive component for Ki67 staining. In the cores stained for p53 the in situ core is negative, with positivity in the invasive core. Vimentin expression is seen in only the stroma and alveolar macrophages in the in situ core, with tumor cell expression identified in the invasive core
Targeted next generation sequencing
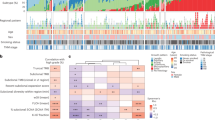
To identify genomic mutations driving invasiveness in early lung adenocarcinomas, 18 C1 tumors were subjected to targeted next generation sequencing of both in situ and invasive areas. We also subjected 5 C2 tumors to sequencing in order to test our hypothesis that in these tumors, in situ growth is in fact facultative surface outgrowth of highly transformed cells with the biological ability to invade. In every case, microscopically identified in situ and invasive epithelial cells were dissected out by LCM prior to panel DNA sequencing. Sequence data are summarized in Fig. 4.
Summary of the variants identified in C1 lesions and those C2 tumors for which DNA was extracted from both in situ and invasive tumor regions. Green represents variants identified in both in situ and invasive regions of the tumor. Red represents variants private to the invasive region. Although cases were tested using a 22-gene next generation sequencing panel, variants in the 23 cases tested were only called in seven of these genes, as represented
Taking the 23 tumors together, mutations in KRAS, EGFR, and BRAF were ubiquitous; one of the genes carried a driver mutation in 74% of this small cohort. In every case, these mutations were shared by both in situ and invasive areas, indicating that they are extremely early events, occurring during the preinvasive stage of tumor development.
The 18 C1 lesions yielded a total of 20 somatic mutations, and only three tumors did not contain detectable mutations. Three tumors had mutations which were private to the invasive portion of the tumor, and which are therefore likely to be driving invasive behavior. Invasion-specific TP53 mutations were present in two of these cases, one of which harbored a second invasion-private mutation in PIK3CA. The third case had an invasion-private mutation in SMAD4. Crucially, no mutations were private to in situ regions.
A total of nine mutations were identified in the five C2 tumors, with all tumors showing at least one mutation. All mutations were shared between in situ and invasive regions.
Even with the limitations of a small sample size the difference in the overall rate of TP53 mutations between the C1 (17%) and C2 tumors (80%) was statistically significant (p = 0.006). Furthermore, two of the three TP53 mutations in C1 lesions were private to the invasive component, so the difference in rate of TP53 mutations in C1 and C2 in situ tumor areas was even greater (6–80%, p = 0.0004). This concurs with the immunohistochemical assay for mutant p53 protein, and further supports the idea that C2-type in situ growth is biologically different from that seen in C1 tumors.
Full details of the variants called are given in Supplementary Data File 2.
For completion all tumors tested by next generation sequencing were also assessed for ALK and ROS1 driver rearrangements. Both assays were negative in all cases tested. One case was not tested due to a lack of residual archival tissue (C1_NGS9), although this case showed an exon 19 EGFR deletion on next generation sequencing analysis.
Prognostic power of Ki-67 in in situ tumor areas
Having observed that in situ/lepidic tumor growth can be of two distinct biological different types, ‘precursor’ (as seen in C1 tumors) and ‘outgrowth’ (as seen in C2), and that these are measurably different in their proliferation rates, we set out to test the hypothesis that precursor type in situ growth is prognostically distinct from invasive type in situ growth in a large set of adenocarcinomas regardless of subtype or stage.
To achieve this, we assessed the Ki67-positive proliferative fraction of tumor cells in tissue microarrays constructed from our entire cohort of 994 adenocarcinoma patients with three cores per patient. Individual cores were morphologically classified as being of in situ or invasive growth pattern. For each case, we calculated a median proliferation score for in situ cores, or for invasive cores, or in cases where both patterns were represented in the microarray, for both areas. Seven hundred and fifty-six of these cases had an invasive pattern core scored for Ki67% and were eligible for survival analysis. One hundred and forty-three of these cases had an in situ pattern core scored for Ki67% and were eligible for survival analysis.
In order to identify a rational cut-off to separate biologically preinvasive tumor from invasive tumor outgrowth we compared proliferation rates in in situ areas from C1 precursor and C2 outgrowth tumors using the focussed tissue microarray data described above (Fig. 5a). A proliferation fraction of around 10% is optimal for separating C1 from C2 in situ growth patterns, so this level was used as a cut-off. An in situ pattern Ki67-positivity rate ≤10% is therefore most likely to represent ‘precursor’ pattern, while a score of ≥10% is more likely to represent ‘outgrowth’ pattern.
a Kernel plot demonstrating the distribution of Ki67 fraction in the in situ component from the LATTICe-A cohort. b Overall survival analysis over 5 years of 756 cases from the entire LATTICe-A cohort with cases classified by the Ki67 fraction in the invasive component of the tumor. c Overall survival analysis over 5 years of 143 cases from the entire LATTICe-A cohort with cases classified by the Ki67 fraction in the in situ component of the tumor
When these values are entered into Kaplan–Meier survival models, proliferation in in situ cores in the tissue microarray is highly prognostic (Fig. 5c). In contrast, the same cut-off applied to proliferation rate in invasive cores is only of borderline significance (Fig. 5b).
This shows that high- and low-proliferative patterns of in situ growth are strongly related to tumor virulence, and we suggest that this is because the ‘outgrowth’ type in situ pattern is likely to reflect nearby invasive disease, whereas ‘precursor’ type growth is relatively innocuous, and may be encountered in entirely preinvasive lesions.
Discussion
A large proportion of resected lung adenocarcinomas contain a proportion of in situ/lepidic growth pattern [17]. The pattern is easily recognized in tissue sections morphologically and is appreciable in CT imaging as ground glass opacity [18]. In both histological and radiological studies, a high proportion of in situ pattern or ground glass opacity is known to predict relatively good outcome clinically [19], as might be expected given that tumor cells in situ are by definition unable to metastasize. For this reason, there is a widespread assumption that in situ tumor proliferation always represents a precursor to invasive disease. However, there has as yet been no systematic investigation of the molecular and/or genomic significance of precisely histologically defined in situ growth pattern.
In this study, we present morphological, immunohistochemical, and genomic evidence that in situ growth in partially invasive adenocarcinomas encompasses two distinct modes of tumor growth (summarised in Table 4). The first is a precursor state, in biological terms a relic from before the acquisition of invasive potential, encountered in early adenocarcinomas as an invasive clone emerges from it, designated in this study as C1 tumors. This is precisely analogous to preinvasive lesions seen in other carcinomas, such as Bowen’s disease in the skin or colonic adenomas. The second is a high-grade outgrowth of biologically advanced malignancy, opportunistically growing on the alveolar surface, in what we designate C2 lesions. This appearance of carcinoma in situ seen ‘creeping’ along an epithelial surface adjacent to high-grade invasive cancer is a phenomenon commonly observed at the periphery of invasive carcinomas including colorectal, squamous, and breast carcinomas. As in other carcinomas, the precursor lesion in lung adenocarcinoma is characteristically of lower proliferation rate and nuclear grade than more highly transformed in situ lesions.
Furthermore, we find that the precursor pattern, as judged by proliferation rate, is associated with better patient outcomes than the outgrowth pattern across a wider cohort of resected adenocarcinomas. That the in situ component Ki67% represents a stronger predictor than the same analysis in the invasive component across all resected adenocarcinomas may reflect ‘outgrowth’ being a marker of aggressive disease in lung adenocarcinoma, regardless of stage and subtype. This distinction is of potential clinical importance.
Whilst it is noted that there is no difference in overall survival difference between C1 and C2 tumors, this may be limited by sample size. Nodal disease was more frequent in C2 tumors and this of course affects overall stage which impacts upon survival, though classification of ≤20 mm adenocarcinomas was performed blinded to nodal status. Ideally only tumors with combined lepidic and acinar growth would have been included in this study, but such restrictive inclusion would have limited this to a far less meaningful cohort of only 20 cases. A small number of tumors which were solid-predominant or showed a pleomorphic component in this cohort were categorized as Noguchi C. These rare tumors may represent the capture, at excision, of a short-lived and transient stage in their evolution, in which the high-grade element has appeared very recently and is poised to overgrow the whole region.
Proportional growth pattern scoring is now a central part of tumor classification under the WHO diagnostic guidelines, and is known to be highly prognostic [20, 21]. The separation of in situ growth into two categories might improve this further. For example, in small biopsies, it is not uncommon only to receive a fragment of tumor displaying in situ growth. Our data suggest that in these circumstances proliferation rate might usefully separate precursor lesions from outgrowth from invasive cancer, with associated prognostic impact. Furthermore, it may be possible to prognostically separate precursor from outgrowth patterns by measurement of metabolic activity by positron emission tomography-CT in ground glass opacities.
Our collection of genomic data was limited in size, both because appropriate lesions are rare, and due to the exacting nature of such careful microdissection. Invasion-private mutations were detected in 3 of 18 C1 tumors sequenced. In the five C2 tumors analyzed, no genomic progression was identified between in situ and invasive elements. The 23 cases sequenced support our model, although definitive conclusions cannot be drawn from a series of this size. The tumors sequenced were skewed toward C1 lesions in order to meet the wider objective of identifying genomic step changes, although a wider sample of C2 tumors and broader sequencing would inform this further. In addition, further analyses such as spatial transcriptomics may more accurately determine whether the morphological step changes seen in C1 tumors is genuine cause of invasive phenotype. While not feasible on this archival series this may be possible in other studies with optimal tumor material.
Driving mutations in five genes were observed in more than one case: KRAS, BRAF, EGFR, TP53, and PIK3CA. KRAS, BRAF, and EGFR mutations were always shared between in situ and invasive areas, implying very early roles in the progression of preinvasive disease, and confirming their status as classic truncal events. This reduplicates the early (i.e., pregenome doubling) timing of these invents as inferred from genomic data in the TRACERx study [7]. We found that TP53 mutations were private to invasive areas in two cases, supporting a wealth of cell biological and mouse model evidence that links TP53 to invasive behavior [4, 6, 22, 23]. However, we also identified an area of in situ proliferation harboring TP53 mutations, in case 11, a mixed-pattern adenocarcinoma. TP53 mutation is not, therefore, sufficient to compel invasive behavior in human lung tissue. Four cases of the series of 23 (17%) showed no somatic mutation. This is in line with a large cohort of lung cancers tested with the same gene panel [24].
The remaining invasion-private driver mutations were in SMAD4 and PIK3CA, implicating a causal role for dysregulation of these pathways in invasive behavior. Activating PIK3CA mutations are not uncommon in lung cancer [25, 26], and have previously been shown to drive invasive behavior in colon cancer cells [27] and to favor epithelial–mesenchymal transition in a mouse model of breast cancer [28], but this is the first direct demonstration of PIK3CA mutation being related to a switch to invasive behavior in human lung cancer. SMAD4 loss, while most well known in the context of gastrointestinal malignancy, is rarely seen in lung adenocarcinomas [7, 24], although it has previously been described as a late event [7].
These data show a very few of the probable plethora of pathways by which natural selection can generate invasive behavior in lung adenocarcinoma, most of which remain undescribed. We think it likely that many of our C1 cases would show evidence of genomic progression via point mutations in other genes, by rearrangements, or by genomic gains and losses. However, it also entirely possible that invasive behavior may arise from preinvasive lesions by dysregulation of epigenetic mechanisms.
References
Ishizumi T, McWilliams A, MacAulay C, et al. Natural history of bronchial preinvasive lesions. Cancer Metastasis Rev. 2010;29:5–14.
Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–85.
Murakami S, Ito H, Tsubokawa N, et al. Prognostic value of the new IASLC/ATS/ERS classification of clinical stage IA lung adenocarcinoma. Lung Cancer. 2015;90:199–204.
Nakanishi H, Matsumoto S, Iwakawa R, et al. Whole genome comparison of allelic imbalance between noninvasive and invasive small-sized lung adenocarcinomas. Cancer Res. 2009;69:1615–23.
Soh J, Toyooka S, Ichihara S, et al. Sequential molecular changes during multistage pathogenesis of small peripheral adenocarcinomas of the lung. J Thorac Oncol. 2008;3:340–7.
Yoo SB, Chung JH, Lee HJ, et al. Epidermal growth factor receptor mutation and p53 overexpression during the multistage progression of small adenocarcinoma of the lung. J Thorac Oncol. 2010;5:964–9.
Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–21.
Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–51.
Yatabe Y, Takahashi T, Mitsudomi T. Epidermal growth factor receptor gene amplification is acquired in association with tumor progression of EGFR-mutated lung cancer. Cancer Res. 2008;68:2106–11.
Murphy SJ, Wigle DA, Lima JF, et al. Genomic rearrangements define lineage relationships between adjacent lepidic and invasive components in lung adenocarcinoma. Cancer Res. 2014;74:3157–67.
Noguchi M, Morikawa A, Kawasaki M, et al. Small adenocarcinoma of the lung. Histologic characteristics and prognosis. Cancer. 1995;75:2844–52.
Nagayoshi Y, Yamamoto K, Hashimoto S, et al. An autopsy case of lepidic pulmonary metastasis from cholangiocarcinoma. Intern Med. 2016;55:2849–53.
Okafuji T, Sakai S, Yoshimitsu S, et al. Pulmonary metastasis from pancreatic cancer: a case showing biphasic radiological and histological patterns. CMIG Extra: Cases. 2004;28:68–71.
Tsao MS, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol. 2007;25:5240–7.
Choi Y, Lee HJ, Jang MH, et al. Epithelial-mesenchymal transition increases during the progression of in situ to invasive basal-like breast cancer. Hum Pathol. 2013;44:2581–9.
Vellinga TT, den Uil S, Rinkes IH, et al. Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene. 2016;35:5263–71.
von der Thüsen JH, Tham YS, Pattenden H, et al. Prognostic significance of predominant histologic pattern and nuclear grade in resected adenocarcinoma of the lung: potential parameters for a grading system. J Thorac Oncol. 2013;8:37–44.
Aokage K, Miyoshi T, Ishii G, et al. Influence of ground glass opacity and the corresponding pathological findings on survival in patients with clinical stage I non-small cell lung cancer. J Thorac Oncol. 2018;13:533–42.
Moon Y, Sung SW, Lee KY, et al. Pure ground-glass opacity on chest computed tomography: predictive factors for invasive adenocarcinoma. J Thorac Dis. 2016;8:1561–70.
Suh YJ, Lee HJ, Kim YT, et al. Added prognostic value of CT characteristics and IASLC/ATS/ERS histologic subtype in surgically resected lung adenocarcinomas. Lung Cancer. 2018;120:130–6.
Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol. 2013;8:52–61.
Lewis BC, Klimstra DS, Socci ND, et al. The absence of p53 promotes metastasis in a novel somatic mouse model for hepatocellular carcinoma. Mol Cell Biol. 2005;25:1228–37.
Wang SP, Wang WL, Chang YL, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694–704.
Moore DA, Balbi K, Ingham A, et al. Analysis of a large cohort of non-small cell lung cancers submitted for somatic variant analysis demonstrates that targeted next-generation sequencing is fit for purpose as a molecular diagnostic assay in routine practice. J Clin Pathol. 2018;71:1001–6.
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–21.
Samuels Y, Diaz LA Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–73.
Yuan W, Stawiski E, Janakiraman V, et al. Conditional activation of Pik3ca(H1047R) in a knock-in mouse model promotes mammary tumorigenesis and emergence of mutations. Oncogene. 2013;32:318–26.
Acknowledgments
This work was supported by a CRUK Center infrastructure award [C1362/A18081] and a Hope Against Cancer Small Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Moore, D.A., Sereno, M., Das, M. et al. In situ growth in early lung adenocarcinoma may represent precursor growth or invasive clone outgrowth—a clinically relevant distinction. Mod Pathol 32, 1095–1105 (2019). https://doi.org/10.1038/s41379-019-0257-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0257-1
This article is cited by
-
The artificial intelligence-based model ANORAK improves histopathological grading of lung adenocarcinoma
Nature Cancer (2024)
-
Evolutionary characterization of lung adenocarcinoma morphology in TRACERx
Nature Medicine (2023)
-
Geospatial immune variability illuminates differential evolution of lung adenocarcinoma
Nature Medicine (2020)