Abstract
Whole slide imaging is Food and Drug Administration-approved for primary diagnosis in the United States of America; however, relatively few pathology departments in the country have fully implemented an enterprise wide digital pathology system enabled for primary diagnosis. Digital pathology has significant potential to transform pathology practice with several published studies documenting some level of diagnostic equivalence between digital and conventional systems. However, whole slide imaging also has significant potential to disrupt pathology practice, due to the differences in efficiency of manipulating digital images vis-à-vis glass slides, and studies on the efficiency of actual digital pathology workload are lacking. Our randomized, equivalency and efficiency study aimed to replicate clinical workflow, comparing conventional microscopy to a complete digital pathology signout using whole slide images, evaluating the equivalency and efficiency of glass slide to whole slide image reporting, reflective of true pathology practice workloads in the clinical setting. All glass slides representing an entire day’s routine clinical signout workload for six different anatomic pathology subspecialties at Memorial Sloan Kettering Cancer Center were scanned on Leica Aperio AT2 at ×40 (0.25 µm/pixel). Integration of whole slide images for each accessioned case is through an interface between the Leica eSlide manager database and the laboratory information system, Cerner CoPathPlus. Pathologists utilized a standard institution computer workstation and viewed whole slide images through an internally developed, vendor agnostic whole slide image viewer, named the “MSK Slide Viewer”. Subspecialized pathologists first reported on glass slides from surgical pathology cases using routine clinical workflow. Glass slides were de-identified, scanned, and re-accessioned in the laboratory information system test environment. After a washout period of 13 weeks, pathologists reported the same clinical workload using whole slide image integrated within the laboratory information system. Intraobserver equivalency metrics included top-line diagnosis, margin status, lymphovascular and/or perineural invasion, pathology stage, and the need to order ancillary testing (i.e., recuts, immunohistochemistry). Turnaround time (efficiency) evaluation was defined by the start of each case when opened in the laboratory information system and when the case was completed for that day (i.e., case sent to signout queue or pending ancillary studies). Eight pathologists participated from the following subspecialties: bone and soft tissue, genitourinary, gastrointestinal, breast, gynecologic, and dermatopathology. Glass slides signouts comprised of 204 cases, encompassing 2091 glass slides; and digital signouts comprised of 199 cases, encompassing 2073 whole slide images. The median whole slide image file size was 1.54 GB; scan time/slide, 6 min 24 s; and scan area 32.1 × 18.52 mm. Overall diagnostic equivalency (e.g., top-line diagnosis) was 99.3% between digital and glass slide signout; however, signout using whole slide images showed a median overall 19% decrease in efficiency per case. No significant difference by reader, subspecialty, or specimen type was identified. Our experience is the most comprehensive study to date and shows high intraobserver whole slide image to glass slide equivalence in reporting of true clinical workflows and workloads. Efficiency needs to improve for digital pathology to gain more traction among pathologists.
Similar content being viewed by others
Introduction
Recent advancements in technology have moved pathology closer towards an enterprise wide digital practice. Innovative technologies have allowed for digitization of glass slides to be rendered, viewed, navigated, and interpreted digitally, comparable to a conventional microscope. The College of American Pathologists [1] has provided guidelines for validation of digital pathology systems and digital pathology has since been approved for primary diagnosis by the Food and Drug Administration (FDA) [2]. Validation of digital pathology systems prior to use in regulated clinical practice and non-clinical environments is necessary [3]. Many studies in the literature show high accuracy for digital pathology with high fidelity in digitizing glass slides and reporting on the whole slide images [4,5,6,7,8,9,10,11,12,13,14,15,16,17]. The major drawback in most prior studies is the utilization of representative slides to demonstrate non-inferiority [18] of the whole slide image, or focusing on select pathology use cases. In addition, in the aforementioned studies, only the overall “top-line” diagnosis was the main endpoint for concordance, as opposed to evaluating the broad range of data elements required for pathology reporting in clinical practice; or the inclusion of other downstream processes (i.e., laboratory information system integration, ordering of ancillary studies, review of prior material, consultation) reflective of actual pathology practice. Another confounding factor in evaluating equivalency and efficiency for whole slide image signout is the large number of digital pathology vendors with varied digital pathology solutions and different needs. The current Food and Drug Administration model does not permit the use of different components to be exchanged with the components of the approved digital pathology system, enabled for primary diagnosis using whole slide images. This flexibility is often demanded by large academic centers with multiple use cases for varied hardware.
Our goal is to compare diagnostic workflows in pathology using whole slide images with that of traditional brightfield microscopy for anatomic pathology in a large academic center. In contrast to prior studies, our study assesses complete pathology workloads with thorough appraisal of variables including various stains (i.e., frozen sections, special stains, immunohistochemistry) and includes the review and reporting of all pathology relevant material received (i.e., current or prior patient slides, radiologic studies, clinical history and notes, etc). The aim of this study includes replicating pathology clinical workflow as a digital pathology signout compared to brightfield microscopy; and to compare equivalency and efficiency of glass slide and whole slide image reporting of true anatomic pathology clinical workloads.
Materials and methods
Case selection
This prospective study included cases from our large academic tertiary comprehensive cancer center. Pathologists’ clinical service days were randomly selected for glass slide signout. Each signout day was composed of all specimen classes including biopsy, surgical resections, and departmental consultation material (e.g., pathology material from other institutions). At our institution trainees preview and compose a preliminary report for each case prior to reviewing with a faculty pathologist for final reporting. For this study, there was no trainee involvement in handling or previewing of the cases. The pathologists directly received the glass slides as distributed from the central laboratory, each case with the respective paper surgical requisition. Each pathologist received their respective case load on their randomly assigned clinical service day, comprised of that day’s entire clinical volume (i.e., biopsies, resection specimens, and consultation) within their subspecialty. Pathologists reviewed an entire day’s workload including all slides and levels taken from biopsies. All slides for all cases were reviewed including immunohistochemical stains and special stains submitted with consultation cases. Cases where frozen sections had been performed were reviewed together with the corresponding frozen section controls as per the pathology department routine workflow. Every effort was taken to directly replicate and mirror the existing pathology workflow in our department. A separate pathologist not participating in case signout validated all cases included in the study. All cases were live clinical cases, such that they were reported by traditional glass microscopy first, prior to digital review after a prolonged washout period.
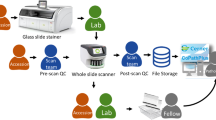
Whole slide scanning protocol
After all cases were reported, the glass slides were retrieved from the pathologists’ offices. All cases were de-identified and re-accessioned in the test environment of the laboratory information system. All clinical documentation, requisitions, and radiologic materials that were available for the initial glass slide signouts were also de-identified and digitally made available in the test environment. All glass slides were de-identified with a non-patient identifiable slide label while keeping all relevant slide label information (i.e., stain type, tissue block, and slide information). Glass slides were accessioned according to each pathologists’ initials as a respective number wheel (i.e., ABC-01) and scanned by Leica Aperio AT2 (Leica Biosystems, Buffalo Grove, Illinois, USA) whole slide scanners at ×40 (0.25 µm/pixel) equivalent magnification using a 20×/0.75NA Plan Apo objective and ×2 automatic optical magnification changer. The pathology laboratory and information technology designation of glass slide label templates have 2D barcodes that interface with Cerner CoPathPlus through the Leica Aperio eSlide manager database. Whole slide images are accessed through the PICSPlus module in CoPathPlus and launched into the MSK Slide Viewer, an internally developed whole slide scanner, vendor agnostic whole slide image viewer.
MSK Slide Viewer
The MSK Slide Viewer is an internally developed, institutional standard whole slide image viewer. As per the institution scanning protocol, prospective clinical cases have been digitally scanned and interfaced with the laboratory information system. For over 1 year, all pathologists at our institution have used the MSK Slide Viewer and reviewed any retrospectively digitized whole slide images for prospective clinical cases. The web-based viewer is initiated within the pathologist’s default web browser. The viewer launches from the PICSPlus module in Cerner CoPathPlus and visualizes any scanned whole slide images for that case (e.g., all, multiple, or individual); and is controlled through the computer mouse and keyboard. For visualization, it includes standard whole slide image viewer software functionality such as thumbnail viewing, slide label viewing, zooming, panning, slide navigation, and co-registration of multiple slides. For annotation, intuitive tools such as a ruler for measurements, tracking of viewed slide regions, screenshots, and comments are integrated. As reinforcement, an additional training session was performed for the pathologists in the study prior to the digital signout sessions.
Glass and digital signout
Our institution follows a subspecialty signout approach where all pathologists are focused on predominantly one or two subspecialties. Pathologists utilized a standard institutional computer workstation configuration (8GB RAM) using a traditional computer mouse as an input device with dual 24″ monitors (screen resolution 1920 × 1200). Administrative staff were assigned for data collection and were present in the pathologists’ office for the duration of both glass and digital signout sessions. Data captured from each signout session included, subspecialty, start and end of each case, specimen type (i.e., biopsy, resection, consultation), part type description (i.e., organ site), number of slides per case, and number of ordered recuts and special stains. Recorded diagnostic information included top-line diagnosis, margin status, lymphovascular and perineural invasion, pT and pN stage. Other data collected included technical difficulties encountered, review of prior patient material, and documentation of interruptions (e.g., instances and duration), if any.
Between glass and digital signouts there was a 13-week washout period. The glass signout session was in the live production laboratory information system, whereas the digital signout was completed in the test environment. The laboratory information system production and test environments mirror each other and have equivalent performance; all reporting tools and functions are available. Pathologists reported all cases according to their routine workflow (i.e., free-text, worksheet templates, synoptic worksheets). Prior patient pathology reports and slides (from prior patient’s pathology) were made available to the pathologists for both signout sessions. If prior pathology slides were not immediately available, the pathologists were required to request glass slides from the pathology department slide archive, similar to the current clinical workflow. For many of the cases prior slides had been scanned into the system previously and these were available as well. Patient medical record numbers were also provided to the administrators for pathologist review of the available clinical application systems (i.e., electronic medical record, radiology picture archiving and communication system (PACS)). A snapshot of the signout day was captured to collect the equivalency data from the glass signout sessions and was compared to the digital signouts. After both signouts, an experience survey was distributed to the pathologists. Technical difficulties were also recorded.
Equivalency
Metrics to evaluate intraobserver equivalency were recorded to reflect the top-line diagnosis, surgical margin status, lymphovascular and/or perineural invasion, pathology tumor and lymph node stage, need for ancillary testing (i.e., recuts, special stains, immunohistochemistry), need for intradepartmental and peer/consensus consultations. For the digital signout, the same equivalency metrics were recorded, with the addition of “did you remember this case” including specifics questions related to which, if any, of the aforementioned metrics were remembered (e.g., remembering the margin being positive or the presence of lymphovascular invasion). Lack of concordance was defined as having a significant impact in clinical management. A pathologist who did not participate as a reader in this study adjudicated concordance of diagnoses.
Efficiency
Efficiency data were defined as the signout turnaround time for each case. The start of each case was defined when the pathologist opened the case in the laboratory information system either by scanning the case barcode or manually entering the accession number and opening the case in the laboratory information system. The end time was stamped when the pathologist completed the case for the day, either by sending the completed case to a signout queue, or if the pathologist closed the case, pending ancillary studies that were ordered at the time of signout. The turnaround times for glass slide and digital signouts were compared.
Results
Eight subspecialized pathologists, referred to hereafter as readers, reported on cases from six subspecialties; with one reader for each, and two for the breast and genitourinary subspecialties. Subspecialties included: breast, bone and soft tissue, dermatopathology, gastrointestinal, genitourinary, and gynecologic pathology (Table 1). Table 1 refers to the slide and case distribution by reader. A total of 2091 glass slides from 204 cases were reviewed and 2073 whole slide images from 199 cases. The readers completed 1075 reads (e.g., diagnoses by part) in total, glass slides, 541; whole slide images, 534. Of the 2091 glass slides that were reviewed in the initial optical signout, 18 slides from five cases were sent out to other institutions due to clinical necessity (e.g., patient being treated elsewhere) prior to being digitized and were excluded from subsequent analysis. Table 2 represents the case distribution by specimen type, of which 136 were in-house cases (e.g., biopsies and resections performed at our institution) and 68 cases sent for consultation from other institutions. There was a mean of six slides per biopsy case (range, 1–42) and 17 per resection case (range, 2–61). Specimens types in this signout dataset included a standard distribution of cases as typically expected in a large tertiary care center with a primary focus on cancer (Table 3).
Technical data
From the 2091 glass slides, there were 2221 glass slide scans, including 148 glass slides that needed to be rescanned (7%). All whole slide images were reviewed by technicians or the referee pathologist (pathologist not participating as a reader) as part of the quality assurance process prior to initiation of primary digital signout. Quality assurance accounted for 80 rescans, where the entire bounds of tissue present on the slide were not scanned and present in the whole slide image. True scanner technical error necessitated 68 whole slide image rescans. Of these 68 rescans, 45 were hematoxylin and eosin stained slides, and 23 were immunohistochemical stained slides. Twenty (87%) of the 23 immunohistochemical stain rescans were immunohistochemical stains that were reported as negative. Four hematoxylin and eosin stained slide rescans were due to barcode recognition malfunctions: these glass slides were scanned and whole slide image files were generated; however, the 2D barcode did not interface with the laboratory information system and would not have been present to launch within the laboratory information system for viewing during signout. All other hematoxylin and eosin and immunohistochemical stains that required rescans were due to focus point errors or incomplete tissue detection (i.e., blurry tiles). Of the 68 total rescans, 64 slides were regarded as the true rescan rate, representing 2.89% of all glass slides. The median scan area was more than double the length of the “industry standard” (e.g., 15 × 15 mm) by most conventional digital pathology hardware vendors, measured at 32.1 mm in length, and 18.5 mm in width. The median scan time per slide at ×40 equivalent resolution (0.25 µm/pixel) was 6 min and 24 s. The median file size was 1.54 gigabytes (GB).
Equivalency
A read is defined in this study as a diagnosis for a specimen part (i.e., for each case, there may be one or multiple parts, comprising of one or many slides). A total of 1075 reads were performed by both glass and whole slide image, 541 and 534, respectively (Table 2). Of the 1064 matched glass slide and whole slide image diagnoses, there were four diagnostic discrepancies, corresponding to a 99.3% glass slide to whole slide image diagnostic concordance rate (530/534 reads). Diagnostic discrepancies are defined as any finding identified by one modality and not identified on the other that would be clinically significant. The four discrepant diagnoses are detailed in Table 4. Three occurred in the genitourinary and one in the breast subspecialties. Size measurements performed by digital annotation were 4% larger than compared to the glass slide measurements. Readers perceived that, when using the digital ruler, they were being more precise with their measurements. Concordance for presence and absence of lymphovascular and/or perineural invasion was overall 83.8% for each part-level diagnosis. However, within parts when lymphovascular and/or perineural invasion were identified as being present, there was only 47% concordance between digital and glass microscopy. The pathology tumor and node stage (e.g., pT, pN) was 97% concordant. Of the four reads that had discrepancies, the one breast read by glass slides showed lobular carcinoma in situ with microinvasion, compared to only lobular carcinoma in situ identified on the whole slide image. Two of the three genitourinary discrepancies were diagnoses not identified in the whole slide image reads, where: in one read of a testis resection, a yolk sac tumor component was not identified in association with teratoma; and the other discrepant read did not identify a secondary Gleason grade pattern of 5 (was read on whole slide image as Gleason patterns 4+3). The third genitourinary discrepant read was a urethral biopsy that was read as non-invasive urothelial carcinoma on whole slide image and benign by glass read. Upon review of the discrepant cases, the diagnoses based on glass slide reads were agreed to be correct. An increase in the need for intradepartmental/peer consultations occurred for two digital cases when compared to cases with glass slides; seven digital and five glass slide cases, respectively, required consultation. All five glass slides cases were included in the seven whole slide image cases where peer consultation was requested. There were no significant differences in the need for prior patient pathology review between digital and glass slide reads. Table 5 summarizes the equivalency evaluation.
Efficiency
The total cumulative glass signout time for all readers was 39:52:02 (hours:minutes:seconds), and the digital signout time was 48:46:51. The total difference of signout turnaround time was 8 h and 52 min, equating to an increase of 1 h and 6 min, per reader signout session. The breakdown of glass and digital signout times comparatively by reader is shown in Table 5. All readers, except one, had a longer digital turnaround time, per complete signout session, compared to glass. Calculated turnaround time per slide shows the median glass signout time was 1 min 13 s compared to 1 min 32 s per whole slide image. This amounts to a median 19 s (26%) increase per slide and 2 min 57 s increase per case when signing out digitally. Each reader had variable intraobserver delta turnaround time comparing their glass to digital signout efficiency. The readers with the fastest and slowest signout turnaround times were dissimilar in comparison to the proportional delta change in time for glass and digital signouts. For example, Pathologist E had the largest delta change in signout time (e.g., 61%), however, that reader had the fastest glass signout time per slide. One reader was faster when reading whole slide image, the other seven readers were faster by traditional glass microscopy. There was no statistically significant difference in efficiency based on the reader, specimen type (i.e., biopsy versus resection), or subspecialty (p > 0.05). Figure 1 shows a visualization of the signout times sorted by specimen type and case signout times normalized by slide (Fig. 1). Collectively, 70 (35%) of the 199 cases had faster signout times by using whole slide image than compared to glass slides. Of the 70 cases, there were 44 cases (35 in-house, 9 consultations) with specimen types as biopsy only and 26 cases (22 in-house, 4 consultations) with resections +/− biopsies. The efficiency per subspecialty and reader showed that breast and bone & soft tissue cases had the slowest diagnostic turnaround times in this study, with dermatopathology having the fastest diagnostic turnaround times.
Comparative efficiency of reporting cases with glass slides and whole slide images. GT glass time, DT digital time, Bx biopsy, C consult, Derm dermatopathology, GI gastrointestinal, GU genitourinary, GYN gynecologic. Note that in most cases, more time was required for one diagnostic modality; the digital slide time (in blue) was longer
Reader recall
After reviewing each digital case, the administrator documented all recall questions during the digital signout session. Ten percentage of the top-line diagnoses were remembered from all cases. In 1.8% of cases, the surgical margin status was remembered. In no cases (0%) was lymphovascular/perineural invasion or pathology stage (e.g., pT, pN) recalled.
Ancillary studies
A total of 629 recuts, special stains, and immunohistochemistry stains were ordered in both signout sessions. Hundred and seventy-seven recuts or levels from the glass and digital signouts were ordered (glass, 105 and digital, 72). Four hundred and fifty-two special or immunohistochemical stains were ordered (glass, 256 and digital, 196). These data show a 31.4% decrease in recuts and 23.4% decrease in special stains/IHC for digital signout. It is unclear whether these decreases are related to case recall or the lack of ancillary studies being performed since the digital reads were all performed after glass signout (i.e., less clinical necessity for confirmatory studies). Of note, Helicobacter pylori immunohistochemical stains were ordered on 100% of the applicable gastric biopsies when reviewed by whole slide image, compared to 33% ordered when reviewing glass slides.
Reader experience survey
A reader experience survey was distributed to all participating readers, of whom six responded (Fig. 2).
Questions in the survey included:
-
1.
Approximately how many years have you been practicing pathology?
-
2.
How many years of experience do you have using digital pathology? (in any capacity)
-
3.
Rate the digital pathology slide viewer.
-
4.
Rate your satisfaction with the launching of slides from within the laboratory information system (CoPath).
-
5.
Rate the quality of the digital slides.
-
6.
Rate your satisfaction with the performance in navigating the digital slides.
-
7.
Would you be interested in testing different input devices (i.e., other than a mouse) to navigate digital slides?
-
8.
How would you rate your overall digital signout experience?
-
9.
How comfortable would you feel signing out digitally, for routine clinical practice?
Of the six survey respondents, the readers in this study have practiced pathology for a median of 15 years (range 6–24). All readers had some experience using digital pathology, with the median number of years being 4.5 years (range 2–10) (Fig. 2a). On a five-point Likert scale (1—very poor, 5—very good), all users rated the MSK Slide Viewer equal to or greater than 3, with a median rating of 4. The reader satisfaction with launching whole slide images from the laboratory information system was ≥4 by all readers. Regarding the overall quality of the whole slide images, all readers reported the experience as ≥4. The satisfaction with the performance in navigating the whole slide images varied among the readers: two responded as very poor, one reader responded with as neutral, and half of the respondents (n = 3) rated the whole slide image navigation as good. Four of the six readers were interested in testing different input devices, to circumvent inefficient digital slide navigation using a conventional computer mouse. The median overall digital signout experience response was rated neutral; one reader each responding as very poor, good, and very good, and three readers responding with neutral. Comfort level regarding the option of signout using whole slide images for primary diagnosis also varied by reader, with the median reader response as uncomfortable. One reader each responded as very uncomfortable, neutral, and comfortable, and three readers responded as uncomfortable (Fig. 2b).
Discussion
As digital pathology matures into a potential future standard of care, institutions must validate internally their intended use. This large preclinical validation study substantiates a distinct paradigm for defining equivalency between the reporting of pathology cases by viewing glass slides using an analog microscope, versus computer monitor-displayed whole slide images and a digital workflow. Equivalency in this setting is meant to reflect all aspects of the clinical care setting pertaining to pathology. All prior concordance studies have viewed accuracy as a surrogate for equivalency; however, the full pathology workflow needs to be considered when such a disruptive technology emerges as a potential standard of care. The College of American Pathologists validation guidelines for using whole slide imaging in the diagnostic setting state that each pathology laboratory planning on implementing whole slide image technology should perform their own validation studies for the respective intended clinical use. The guidelines also suggest establishing an intraobserver variability rate and do not recommend any particular order for the reporting of the glass slides or whole slide images [1]. In keeping with these guidelines, our validation study encompassed surgical pathology at our institution, including frozen section tissue slides, special stains, and immunohistochemical studies. We also validated our complete workflow with regard to accessioning of digital cases in the laboratory information system, scanning of whole slide images, launching of whole slide images from within the laboratory information system (e.g., laboratory information system-centric workflow), ordering of ancillary studies through the laboratory information system, and reporting of not only the top-line diagnosis, but of all other pathology parameters. A Q-probe study performed by the College of American Pathologists demonstrated median laboratory discrepancy frequencies of 5.1%, for anatomic pathology [19]. In addition, as previously published and accepted by the Food and Drug Administration, the major discordance rate with a reference standard diagnosis was 4.9% for whole slide images and 4.6% using glass slides [18]. Therefore, in our study, the reference standard was designated as optical microscopy. This study also made efforts to obviate selection and recall bias by randomly selecting clinical service days to include all consecutive cases and specimen types from the signout day. Each case was validated and adjudicated by a separate pathologist not participating in case signout.
A meta-analysis review of the prior concordance studies, including crossover, retrospective, and prospective studies, have demonstrated an upward increase in overall diagnostic concordance over the years of publications [4,5,6,7,8,9,10,11,12,13,14,15,16,17,18, 20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47]. This may be due to a combination of enhanced hardware and software of whole slide imaging systems, or the increased familiarity and confidence of the pathologists using these systems over time. Of all prior studies, the diagnostic intraobserver reported concordance ranged from 63 to 100%, with the mean percentage of diagnostic concordance being 92.4%. For studies encompassing multiple subspecialty disciplines, the range of diagnostic concordance was 75–97%. Our study demonstrates similar results to the prior studies reflecting the high fidelity of the whole slide images to the glass slides, such that the whole slide images reflect accurate representations of the glass slides.
The advantages of whole slide images are not limited to having accurate representation of the glass slides. In order to become a new standard of care, digital pathology systems must embed themselves in current pathology workflows. Although there has been exhaustive literature on the high diagnostic concordance of whole slide images to glass slides, only a few studies have published efficiency rates in a clinical setting. Three studies prior to 2014 reported similar results, such that all diagnoses rendered using whole slide images had a longer time compared to reviewing glass slides [48,49,50]. The average times for whole slide image review and glass slide review were reported as fast as 38 s and 34 s for two whole slide image modalities compared to 23 s for glass slides in one study; and up to 91.9 and 57.1 s by whole slide images and glass slides, respectively, in a second study [48, 49]. Another study reviewing renal transplant biopsies (reviewing H&E and PAS stained slides for each case) showed an average 1.4 times longer to review the whole slide images than the glass slides, at 15.3 and 10.8 min, respectively [50]. These studies published prior to 2014 may not be applicable to the hardware and software standards that are currently available for clinical use. Four additional studies have been published after 2014 and include comparative review of efficiency when comparing whole slide images to glass slides [51,52,53,54]. Randell et al. reviewed slides from three pathology subspecialties (e.g., breast, gastrointestinal, and gynecologic) using two high resolution monitors and an internally developed whole slide image viewer. Twelve pathologists reviewed one oncologic case using both modalities, and no significant difference in total review time was identified between each reader or subspecialty. The mean time to review each slide was 35.7 s per whole slide image and 37.5 s per each glass slide review. Interestingly, the authors noted the time for physical loading and unloading of glass slides on the microscope accumulated 16% of time on the microscope review [51]. Thrall et al. reviewed diagnostic assessment for 752 cases in surgical pathology (including frozen sections, special stains, and IHC), as part of validating multiple whole slide scanners for deployment of a digital pathology system. The overall mean viewing time for whole slide images was 54 s longer than glass slides, however, the authors noted potential confounding factors regarding network streaming delays or server errors. The authors showed a 30% increase in time spent per case by whole slide image review, 235 s versus 181 s [52]. A comparative study of 400 consecutive cases by a single pathologist evaluating general surgical pathology (including biopsies and resections) and cytology, with a fully integrated laboratory information system and digital pathology system, measured turnaround time from the point slides were made available to the point a diagnosis was made or additional investigations were deemed necessary. These times included slide evaluation as well as nondiagnostic time (e.g., dictation, accessing clinical information, and previous history review). The total digital diagnostic time was shorter for whole slide images than for glass slides, 1841 min versus 1956 min. This would suggest having a fully integrated laboratory information management system would provide a total decrease in time to diagnosis (i.e., from case accession to final report) through decreased physical handling of glass slides, less matching of glass slides to requisitions, and with the benefits of a digital workflow [53]. Mills et al. reported on digital diagnostic reporting time of select surgical pathology slides from 510 cases, including biopsy and resection specimens. Assessment times excluded the time to report the diagnosis. The authors concluded that there was a 4 s per case median increase (5.3 s mean increase) in digital assessment time compared to glass slides. Of the three readers, there was significant variability, with the slowest reader having an average of 9.1 more seconds to read a case comprising of whole slide images, and the fastest reader had a 1.2 s mean digital time difference. The authors also illustrated a digital reading effect where readers shortened their digital assessment times with increased experience. The study did not report any significant difference between subspecialty or specimen type pertaining to differences in diagnostic assessment times [54]. Table 6 refers to a summary of the published literature. Our study showed a decrease in digital signout efficiency by a median 19% per case. Of the 70 cases signed out faster when reviewing whole slide images, there were 44 biopsy cases and 26 resections, suggesting whole slide images may be more efficient for cases with less tissue or fewer slides. This may be due to the previously reported conclusions of the time spent to physically remove and place the glass slides on the stage, compared to clicking the subsequent digital thumbnail to load the whole slide image, or having the ability for co-registration, which allows multiple IHC to be synchronously visible at one time. However, no statistically significant differences were noted by reader, subspecialty, or specimen type in our study. The difference could also be related to the number of pathologic parameters included in our study to assess equivalence which is a more accurate reflection of what a pathologist needs to include in a pathology report.
Operational validation metrics
This validation study provides additional important operational metrics for consideration when implementing a digital pathology system. The industry standard area determination upon which whole slide scanner scan times is based is 15 × 15 mm. Based upon scanning 2073 glass slides, the median tissue scan area in routine practice was 32.1 × 18.52 mm, which is 2.6× larger. This reflects scanning of all tissue on the slide, such that if multiple tissue levels were present on a slide, they would be captured. It is reasonable to expect a mix of specimen types at any reasonably sized medical institution, including our large academic center focused on cancer care. When deploying digital pathology systems, validation of scan times based on realistic tissue areas is recommended to ensure appropriate clinical turnaround time, especially for primary diagnosis workflow planning. Digital file scan times and storage sizes for ×40 equivalent resolution (0.25 µm/pixel) resulted in a median scan time of 6 min 24 s and 1.54 GB per slide. We chose to scan all slides at ×40 in order to replicate the availability of this magnification on all optical microscopes used by the reviewers. Whether this level of magnification is a true necessity remains to be evaluated. Of note, our study showed a decrease in ordering of recuts, special stains, and immunohistochemistry in the digital signout session. Although the digital cases were treated as clinical cases, the subsequent reporting of all cases by whole slide images in the test laboratory information system for non-clinical purposes may have biased these data points, and subsequent crossover studies would be of interest.
Limitations/considerations
As of the date of this writing, only one digital pathology system had been approved by the Food and Drug Administration for primary diagnosis in the United States. In this study, our hardware and software were not approved for the intended use of primary diagnosis by the Food and Drug Administration. Thus, all of the cases were reported first by reviewing the glass slides and then subsequently with digital whole slide images. At our institution, there is a high volume and variety of cases, including rare cases that are not routine in practice. Although pathologists may recall one particular aspect of a case, these data suggest that other diagnostic parameters are not easily recalled. The ground truth reference diagnosis was reliant on the optical diagnosis, and each subspecialty dataset was reviewed providing intraobserver concordance. Similar to prior studies, the systems utilized in this study provided high intraobserver diagnostic concordance.
Additionally, this study does not evaluate a digital pathology learning effect, the cases included in this study were gathered from a random signout day, and varied in specimen class and complexity. The pathologist involved in this study also had varied prior experience with digital pathology, although all had an appropriate level of experience with the whole slide image viewer utilized in the study. Efficiency evaluation in this study did not include metrics of future technological improvements, such as new input devices, differences compared to varied monitor resolution, image analysis, machine learning, or computer-assisted diagnosis in relation to productivity. Another potential area to improve whole slide image efficiency not reported in this study is the institutional network. Optimization of network performance is critical in order to deliver appropriate bandwidth and network connectivity. Navigation of whole slide images are dependent on server storage and network performance to remediate delayed loading or rendering of tiles to the pathologists reviewing the whole slide image. Whole slide viewer software performance should also be optimized for appropriate digital clinical workflow. If indicated, launching and loading of numerous slides from the laboratory information system to a separate viewer software should be included in digital pathology system validation to ensure efficient loading times and decrease pathologist frustration. Navigation of whole slide image will also require improvements in hardware or software. Innovative input devices should be sought and tested to efficiently navigate whole slide images. Enhanced software features can enable quicker navigation of whole slide images, rather than click-and-drag motions with a conventional computer mouse. One such study identified the 6-degrees of freedom navigator (Space Mouse Pro, 3DConnexion) to be most preferred input device by the evaluating pathologists [55]. Another means to document improved digital signout efficiency is to investigate turnaround time from the perspective of the entire specimen workflow within pathology, examining from the time of specimen receipt to final diagnosis reporting. This approach has the added benefit of capturing the benefits of a digital workflow on the entirety of the laboratory process (e.g., automated case assembly and distribution), which was not measured in the current study. Assessment of digital pathology workstation ergonomics would also be of value to ensure pathologists’ change in practice will not be compromised when transitioning to a digital workflow.
Future directions
An extended evaluation of all surgical pathology subspecialties is planned as a follow-up to this study and will include primary endpoints of evaluation and validation of interobserver concordance comparing whole slide image to glass slide signout sessions. Future studies to address potential differences in efficiency using variable input devices will be useful to show clinical utility of additional hardware potentially required for successful digital pathology system deployment; evaluating differences in reporting time, including viewer software performance, and user experience, would also be useful. Optimization of network and software performance should also play a role in decreasing the overall median time of reporting whole slide images compared to glass slides. Emerging trends in image analysis and machine learning can be leveraged to enhance capabilities currently unavailable with the analog microscope. Computer-assisted diagnosis and workflow enhancements (such as automated quantification of immunohistochemistry [56, 57] and automated recognition of potential regions of interest such as minimal nodal involvement by metastatic disease) could improve digital signout efficiencies. These computational tools in pathology and better systems to handle whole slide image workflow could be leveraged to triage specimens through automated pre-screening, seamless consultation, and objective biomarker quantification. Such value-added functionalities should only enhance the digital signout experience and further justify the push towards more widespread adoption of digital pathology for primary diagnosis.
Conclusions
This study is the most comprehensive to date and evaluates true anatomic pathology clinical workflow and workload. Our results demonstrate high intraobserver whole slide image to glass slide equivalence in reporting of true clinical workflow and workload as evidenced by diagnosis, 99.3%; grade, 94.1%; stage (pT, pN), >97%; margin status, 100%; and lymphovascular/perineural invasion, 83.8%. Digital pathology reporting of whole slide images showed a 19 s decrease in efficiency per slide and median overall 19% decrease in efficiency per case, 2 min 57 s. Proper validation should include true load testing of scanner throughput with slides generated from the validating laboratory, with subsequent evaluation of digital storage accommodations and network infrastructure. This investigation serves to further validate whole slide images being non-inferior to glass slides from the standpoint of diagnostic concordance, but importantly demonstrates loss of efficiency in the diagnostic turnaround time in a true clinical environment, requiring improvements in other aspects of the pathology workflow to support full adoption of digital pathology.
References
Pantanowitz L, Sinard JH, Henricks WH, et al. Validating whole slide imaging for diagnostic purposes in pathology. Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137:1710–22.
U.S. Food and Drug Administration. FDA News Release: FDA allows marketing of first whole slide imaging system for digital pathology. https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm553358.htm. [Accessed 12 April 2017].
Hanna MG, Pantanowitz L, Evans AJ. Overview of contemporary guidelines in digital pathology: what is available in 2015 and what still needs to be addressed? J Clin Pathol. 2015;68:499–505.
Al-Janabi S, Huisman A, Nap M, et al. Whole slide images as a platform for initial diagnostics in histopathology in a medium-sized routine laboratory. J Clin Pathol. 2012;65:1107–11.
Bauer TW, Schoenfield L, Slaw RJ, et al. Validation of whole slide imaging for primary diagnosis in surgical pathology. Arch Pathol Lab Med. 2013;137:518–24.
Brunelli M, Beccari S, Colombari R, et al. iPathology cockpit diagnostic station: validation according to College of American Pathologists Pathology and Laboratory Quality Center recommendation at the Hospital Trust and University of Verona. Diagn Pathol. 2014;9:S12.
Buck TP, Dilorio R, Havrilla L, et al. Validation of a whole slide imaging system for primary diagnosis in surgical pathology: a community hospital experience. J Pathol Inform. 2014;1:43.
Campbell WS, Lele SM, West WW, et al. Concordance between whole slide imaging and light microscopy for routine surgical pathology. Hum Pathol. 2012;43:1739–44.
Cheng CL, Azhar R, Sng SH, et al. Enabling digital pathology in the diagnostic setting: navigating through the implementation journey in an academic medical centre. J Clin Pathol. 2016;69:784–92.
Fonyad L, Krenac T, Nagy P, et al. Validation of diagnostic accuracy using digital slides in routine histopathology. Diagn Pathol. 2012;7:35.
Gilbertson JR, Ho J, Anthony L, et al. Primary histologic diagnosis using automated whole slide imaging: a validation study. BMC Clin Pathol. 2006;6:4–19.
Goacher E, Randell R, Williams B, et al. The diagnostic concordance of whole slide imaging and light microscopy. Arch Pathol Lab Med. 2017;141:151–61.
Houghton JP, Ervine AJ, Kenny SL, et al. Concordance between digital pathology and light microscopy in general surgical pathology: a pilot study of 100 cases. J Clin Pathol. 2014;67:1052–5.
Jukic DM, Drogowski LM, Martina J, et al. Clinical examination and validation of primary diagnosis in anatomic pathology using whole slide digital images. Arch Pathol Lab Med. 2011;135:372–8.
Mukhopadhyay S, Feldman MD, Abels E, Ashfaq R, Beltaifa S, Cacciabeve NG, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: a multicenter blinded randomized noninferiority study of 1992 cases (Pivotal study). Am J Surg Pathol. 2018;42:39–52.
Pagni F, Bono F, Di Bella C, et al. Virtual surgical pathology in underdeveloped countries: the Zambia project. Arch Pathol Lab Med. 2011;135:215–9.
Snead DR, Tsang YW, Meskiri A, et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology. 2016;68:1063–72.
Tabata K, Mori I, Sasaki T, et al. Whole slide imaging at primary pathological diagnosis: validation of whole slide imaging-based primary pathological diagnosis at twelve Japanese academic institutes. Pathol Int. 2017;67:547–54.
Raab SS, Nakhleh RE, Ruby SG. Patient safety in anatomic pathology. Measuring discrepancy frequencies and causes. Arch Pathol Lab Med. 2005;129:459–66.
Al Habeeb A, Evans A, Ghazarian D. Virtual microscopy using whole slide imaging as an enabler for teledermatopathology: a paired consultant validation study. J Pathol Inform. 2012;3:2.
Al-Janabi S, Huisman A, Jonges GN, ten Kate FJW, Goldschmeding R, van Diest PJ. Whole slide images for primary diagnostics of urinary system pathology: a feasibility study. J Ren Inj Prev. 2014;3:91–6.
Al-Janabi S, Huisman A, Nikkels PG, ten Kate FJ, van Diest PJ. Whole slide images for primary diagnostics of paediatric pathology specimens: a feasibility study. J Clin Pathol. 2013;66:218–23.
Al-Janabi S, Huisman A, Vink A, et al. Whole slide images for primary diagnostics in dermatopathology: a feasibility study. J Clin Pathol. 2012;65:152–8.
Al-Janabi S, Huisman A, Vink A, et al. Whole slide images for primary diagnostics of gastrointestinal tract pathology: a feasibility study. Hum Pathol. 2012;43:702–7.
Al-Janabi S, Huisman A, Willems SM, Van Diest PJ. Digital slide images for primary diagnostics in breast pathology: a feasibility study. Hum Pathol. 2012;43:2318–25.
Arnold MA, Chenever E, Baker PB, et al. The College of American Pathologists guidelines for whole slide imaging validation are feasible for pediatric pathology: a pediatric pathology practice experience. Pediatr Dev Pathol. 2015;18:109–16.
Bauer TW, Schoenfield L, Slaw RJ, Yerian L, Sun Z, Henricks WH. Validation of whole slide imaging for primary diagnosis in surgical pathology. Arch Pathol Lab Med. 2013;137:518–24.
Campbell WS, Hinrichs SH, Lele SM, et al. Whole slide imaging diagnostic concordance with light microscopy for breast needle biopsies. Hum Pathol. 2014;45:1713–21.
Chargari C, Comperat E, Magné N, et al. Prostate needle biopsy examination by means of virtual microscopy. Pathol Res Pract. 2011;207:366–9.
Gage JC, Joste N, Ronnett BM, et al. A comparison of cervical histopathology variability using whole slide digitized images versus glass slides: experience with a statewide registry. Hum Pathol. 2013;44:2542–8.
Gui D, Cortina G, Naini B, et al. Diagnosis of dysplasia in upper gastro-intestinal tract biopsies through digital microscopy. J Pathol Inform. 2012;3:27.
Ho J, Parwani AV, Jukic DM, Yagi Y, Anthony L, Gilbertson JR. Use of whole slide imaging in surgical pathology quality assurance: design and pilot validation studies. Hum Pathol. 2006;37:322–31.
Jen KY, Olson JL, Brodsky S, Zhou XJ, Nadasdy T, Laszik ZG. Reliability of whole slide images as a diagnostic modality for renal allograft biopsies. Hum Pathol. 2013;44:888–94.
Krishnamurthy S, Mathews K, McClure S, et al. Multi-institutional comparison of whole slide digital imaging and optical microscopy for interpretation of hematoxylin-eosin–stained breast tissue sections. Arch Pathol Lab Med. 2013;137:1733–9.
Mooney E, Hood AF, Lampros J, Kempf W, Jemec GB. Comparative diagnostic accuracy in virtual dermatopathology. Skin Res Technol. 2011;17:251–5.
Nielsen PS, Lindebjerg J, Rasmussen J, Starklint H, Waldstrom M, Nielsen B. Virtual microscopy: an evaluation of its validity and diagnostic performance in routine histologic diagnosis of skin tumors. Hum Pathol. 2010;41:1770–6.
Ordi J, Castillo P, Saco A, et al. Validation of whole slide imaging in the primary diagnosis of gynaecological pathology in a university hospital. J Clin Pathol. 2015;68:33–9.
Ozluk Y, Blanco PL, Mengel M, Solez K, Halloran PF, Sis B. Superiority of virtual microscopy versus light microscopy in transplantation pathology. Clin Transplant. 2012;26:336–44.
Reyes C, Ikpatt OF, Nadji M, Cote RJ. Intra-observer reproducibility of whole slide imaging for the primary diagnosis of breast needle biopsies. J Pathol Inform. 2014;5:5.
Risio M, Bussolati G, Senore C, et al. Virtual microscopy for histology quality assurance of screen-detected polyps. J Clin Pathol. 2010;63:916–20.
Rodriguez-Urrego PA, Cronin AM, Al-Ahmadie HA, et al. Interobserver and intraobserver reproducibility in digital and routine microscopic assessment of prostate needle biopsies. Hum Pathol. 2011;42:68–74.
Sanders DS, Grabsch H, Harrison R, et al. AspECT Trial Management Group and Trial Principal Investigators. Comparing virtual with conventional microscopy for the consensus diagnosis of Barrett's neoplasia in the AspECT Barrett's chemoprevention trial pathology audit. Histopathology. 2012;61:795–800.
Shaw EC, Hanby AM, Wheeler K, et al. Observer agreement comparing the use of virtual slides with glass slides in the pathology review component of the POSH breast cancer cohort study. J Clin Pathol. 2012;65:403–8.
van der Post RS, JAWM vanderLaak, Sturm B, et al. The evaluation of colon biopsies using virtual microscopy is reliable. Histopathology. 2013;63:114–21.
Velez N, Jukic D, Ho J. Evaluation of 2 whole slide imaging applications in dermatopathology. Hum Pathol. 2008;39:1341–9.
Wendum D, Lacombe K, Chevallier M, et al. Histological scoring of fibrosis and activity in HIV-chronic hepatitis B related liver disease: performance of the METAVIR score assessed on virtual slides. J Clin Pathol. 2009;62:361–3.
Wilbur DC, Madi K, Colvin RB, et al. Whole slide imaging digital pathology as a platform for teleconsultation: a pilot study using paired subspecialist correlations. Arch Pathol Lab Med. 2009;133:1949–53.
Gui D, Cortina G, Naini B, et al. Diagnosis of dysplasia in upper gastro-intestinal tract biopsies through digital microscopy. J Pathol Inform. 2012;3:27.
Jen KY, Olson JL, Brodsky S, Zhou XJ, Nadasdy T, Laszik ZG. Reliability of whole slide images as a diagnostic modality for renal allograft biopsies. Hum Pathol. 2013;44:888–94.
Velez N, Jukic D, Ho J. Evaluation of 2 whole slide imaging applications in dermatopathology. Hum Pathol. 2008;39:1341–9.
Randell R, Ruddle RA, Thomas RG, Mello-Thoms C, Treanor D. Diagnosis of major cancer resection specimens with virtual slides: impact of a novel digital pathology workstation. Hum Pathol. 2014;45:2101–6.
Thrall MJ, Wimmer JL, Schwartz MR. Validation of multiple whole slide imaging scanners based on the guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2015;139:656–64.
Vodovnik A. Diagnostic time in digital pathology: a comparative study on 400 cases. J Pathol Inform. 2016;7:4.
Mills AM, Gradecki SE, Horton BJ, et al. Diagnostic efficiency in digital pathology. A comparison of optical vs. digital assessment in 510 surgical pathology cases. Am J Surg Pathol. 2018;42:53–9.
Molin J, Lundström C, Fjeld M. A comparative study of input devices for digital slide navigation. J Pathol Inform. 2015;6:7.
Pantanowitz L, Sinard JH, Henricks WH, et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the college of American pathologists pathology and laboratory quality center. Arch Pathol Lab Med. 2013;137:1710–22.
Fine JL. 21st century workflow: a proposal. J Pathol Inform. 2014;5:44.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest related to published content in this article other than listed as follows: TF is co-founder, equity holder and Chief Scientific Officer of Paige.AI; DK is co-founder, equity holder, and consultant of Paige.AI; PJS is a Senior AI Scientist and equity holder of Paige.AI.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hanna, M.G., Reuter, V.E., Hameed, M.R. et al. Whole slide imaging equivalency and efficiency study: experience at a large academic center. Mod Pathol 32, 916–928 (2019). https://doi.org/10.1038/s41379-019-0205-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0205-0
This article is cited by
-
Künstliche Intelligenz in der Pathologie – wie, wo und warum?
Die Pathologie (2024)
-
Digitalisierung der histopathologischen Routinediagnostik
Die Pathologie (2024)
-
Expectations and Experiences Among Clinical Staff Regarding Implementation of Digital Pathology: A Qualitative Study at Two Departments of Pathology
Journal of Imaging Informatics in Medicine (2024)
-
Learning fast and fine-grained detection of amyloid neuropathologies from coarse-grained expert labels
Communications Biology (2023)
-
Screening adequacy of unstained thyroid fine needle aspiration samples using a deep learning-based classifier
Scientific Reports (2023)