Abstract
Amplification of vascular endothelial growth factor A (VEGFA) has been recently reported in TFEB-amplified renal cell carcinomas regardless the level of TFEB amplification. We sought to determine VEGFA amplification by fluorescent in situ hybridization (FISH) and VEGFA mRNA expression by in situ hybridization (RNAscope 2.5) in a series of 10 renal cell carcinomas with TFEB gene alterations, either amplification and/or rearrangement (t(6;11) renal cell carcinoma). TFEB gene rearrangement was demonstrated in eight cases, whereas the remaining two cases showed a high level of TFEB (> 10 copies of fluorescent signals) gene amplification without evidence of rearrangement. Among the eight t(6;11) renal cell carcinomas (TFEB-rearranged cases), one case displayed a high level of TFEB gene amplification and two showed increased TFEB gene copy number (3–4 copies of fluorescent signals). Those three cases behaved aggressively. By FISH, VEGFA was amplified in all three cases with TFEB amplification and increased VEGFA gene copy number was observed in the two aggressive cases t(6;11) renal cell carcinomas with an overlapping increased number of TFEB fluorescent signals. Overall, VEGFA mRNA expression was observed in 8 of 10 cases (80%); of these 8 cases, 3 cases showed high-level TFEB amplification, one case showed TFEB rearrangement with increased TFEB gene copy number, whereas four showed TFEB gene rearrangement without increased copy number. In summary, VEGFA amplification/increased gene copy number and VEGFA mRNA expression occur in TFEB-amplified renal cell carcinoma, but also in a subset of t(6;11) renal cell carcinoma demonstrating aggressive behavior, and in unamplified conventional t(6;11) renal cell carcinoma suggesting VEGFA as potential therapeutic target in these neoplasms even in the absence of TFEB amplification. We finally propose that all the renal tumors showing morphological characteristics suggesting t(6;11) renal cell carcinoma and all unclassified renal cell carcinomas, either high grade or low grade, should immunohistochemically be evaluated for cathepsin K and/or Melan-A and if one of them is positive, tested for TFEB gene alteration and VEGFA gene amplification.
Similar content being viewed by others
Introduction
Molecular classification of renal cell carcinoma has been evolving in the last decades, with the emergence of new entities and new genetic characteristics. To date, MiT family translocation renal cell carcinoma, either involving TFE3 or TFEB gene, has been broadly studied [1] and recent work has lead to the delineation of TFEB-amplified renal cell carcinoma [2,3,4,5,6,7,8]. TFEB amplification in renal cell carcinoma can occur independently of or in association with TFEB rearrangement. TFEB gene rearrangement via chromosome translocation or amplification causes intact TFEB overexpression and drives subsequent expression of immunohistochemical markers such as cathepsin K, Melan-A and HMB45 [9]. However, TFEB-amplified renal cell carcinomas differ from TFEB translocation renal cell carcinomas in several ways [2]. First, they typically occur in older patients (mean 65 years) compared with unamplified TFEB translocation RCC (mean age 31 years). Second, their morphology is usually high grade and less distinctive than the biphasic appearance of the typical TFEB translocation renal cell carcinoma. Third, melanocytic marker expression is less consistent: while all cases have expressed Melan-A, only approximately 50% express cathepsin K and HMB45. Fourth, TFEB-amplified renal cell carcinomas typically have an aggressive clinical course while TFEB translocation renal cell carcinoma usually are indolent.
TFEB gene is located in the short arm of chromosome 6, specifically in the 6p21-p23 region, immediately adjacent to vascular endothelial growth factor A (VEGFA) gene. Given the proximity of those two genes, it has been hypothesized and demonstrated that some renal cell carcinomas showing TFEB amplification harbor concurrent VEGFA amplification [4]. Because amplification of VEGFA has been not investigated in t(6;11) translocation renal cell carcinomas unassociated with TFEB amplification, we analyzed a series of renal cell carcinomas with TFEB gene alterations, either amplification or rearrangement (t(6;11) translocation renal cell carcinoma) to evaluate the occurrence of VEGFA gene copy number and mRNA VEGFA expression.
Materials and methods
Patients and samples
Eight t(6;11) renal cell carcinomas and two unclassified renal cell carcinoma with TFEB gene amplification were retrieved from the files of participating institutions. Seven cases (from case 1 to case 7) have been previously reported and three unpublished cases have been added. All slides were reviewed by two authors (AC and GM). The morphology and immunoprofile of the three additional tumors was recorded.
Immunohistochemistry
Sections from tissue blocks of t(6;11) renal cell carcinomas and TFEB-amplified renal cell carcinoma were immunohistochemically stained with the following antibodies: PAX8 (clone BC12, DSB), cathepsin K (clone 3F9, dilution 1:2000, Abcam), HMB45 (dilution 1:30, Dako), Melan-A (clone A103, dilution 1:50, Novocastra), cytokeratin 8–18 (clone 5D3, dilution 1:100, Novocastra) and CD68 (clone PG-M1, dilution 1:50, Dako). All samples were processed using a sensitive “Bond Polymer Refine” detection system in an automated Bond immunohistochemistry instrument (Leica Biosystems). The appropriate positive and negative controls were concurrently carried out. Labeling for each marker was recorded as the percentage of positive cells.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) was carried out on the 10 carcinomas with TFEB gene alteration using dual color break apart TFEB, TFE3 probe (Cytotest Inc., Rockville, MD, USA) and VEGFA (ZytoVision, Bremerhaven, Germany) probe. Centromeric alpha-satellite specific for chromosome 6 (CEP6) was used as control probes (Vysis-Abbott, Olympus, Rome, Italy) on serial tissue sections. Briefly, 3 µm sections were cut from formalin-fixed paraffin-embedded tissue blocks and mounted on positively charged slides. The slides were dried for 1 h at 60 °C then deparaffinized, rehydrated and fixed in methanol/acetic acid 3:1 for 5 min. Pretreatment was performed at 85 °C for 30 min with 0.1 citrate buffer (pH6) solution followed by pepsin (4 mg/ml in 0.9% NaCl, pH 1.5) treatment for 8 min at 37 °C. After washing and dehydration, 10 µl probe was applied on selected area and sealed with rubber cement. Denaturation was assessed by incubating the slides at 80 °C for 10 min in a humidified atmosphere (Thermobrite System) followed by hybridization overnight at 37 °C. The rubber cement and the cover slip were removed and the slides were washed in 2X SSC/0.3% NP40 for 15 min at room temperature and then at 72 °C for 2 min. Next, the tissue sections were counterstained with 4,6-diamidino-2-phenylindole antifade (Prolong Gold Antifade Reagent Life Technologies) and examined under an X60–X100 oil immersion objective using an Olympus BX61 fluorescence microscope equipped with filters that visualize the different wavelengths of the fluorescent probe.
Scoring was performed by two experienced pathologists (AC and MB). At least 100 neoplastic non-overlapping nuclei were included in the scoring. Ratio between mean copy number of TFEB gene/mean copy number of control centromeric probes CEP6 was ultimately scored. Amplification was defined by the presence of > 10 TFEB/VEGFA fluorescent signals or the locus specific identifier/centromeric alpha-satellite specific probe (LSI/CEP) ratio was ≥ 2.
mRNA in situ hybridization (RNAscope)
The samples were analyzed with RNAscope assay (Advanced Cell Diagnostics, Newark, CA, USA) using RNAscope 2.5 HD Assay-Brown kit and the Probes-Hs-VEGFA. Ten renal tumors (three clear cell renal cell carcinomas, three papillary renal cell carcinomas, two chromophobe renal cell carcinomas and two oncocytomas) were used as control cases. The procedure was performed manually following the manufacturer’s instructions. We used freshly cut 3 µm formalin-fixed and paraffin-embedded slides dried for 1 h at 60 °C. The sections were deparaffinized and treated with the peroxidase block solution for 10 min at room temperature and then with retrieval solution for 15 min at 99 °C. For each case, three sections with targeted probes were incubated: VEGFA, Bacillus subtilis dihydrodipicolinate reductase (DAPB) as negative control and Ubiquitin C (UBC) as positive control. The hybridization was performed for 2 h at 40 °C. Slides were then washed and incubated with the signal amplification solution: amp1 for 30 min at 40 °C, amp2 for 15 min at 40 °C, amp3 for 30 min at 40 °C, amp4 for 15 min at 40 °C, amp5 for 30 min at room temperature, amp6 for 15 min at room temperature and finally with Diaminobenzidine (DAB) for 10 min and hematoxylin for the counterstaining. The results were examined under a standard bright-field microscope at ×60 magnification.
Scoring was performed according to ACD guideline for semiquantitative assessment of RNAscope staining intensity as (0, 1, 2, 3, 4) (https://acdbio.com/technical-support/solutions). A positive result was considered when the neoplastic cells showed 3 or 4 intensity staining.
Results
Clinical characteristics
The clinical features of the 10 patients are detailed in Table 1. Five patients were female and five male (F:M ratio, 1:1). The patients’ ages at diagnosis ranged from 19 to 80 years (mean 45, median 41). The clinical history of patient 1 to patient 7 has been previously reported [8]. Patient 8, HCV infected, initially presented to the emergency department complaining abdominal pain. He underwent a computed tomography (CT) scan, a renal mass was discovered and he underwent radical nephrectomy. Patients 9 and 10 suffered from abdominal pain due to lithiasis of upper urinary tract. In both cases, the renal mass was an incidental finding (Fig. 1) and both patients were treated by partial nephrectomy. Follow-up was available for all patients, ranging from 14 to 48 months (mean 34, median 33). Patients 6 and 7 developed metastasis, as previously detailed. Patient 8 recurred with multiple nodules adjacent to the pancreatic tail, in the perinephric fat infiltrating the psoas muscle, and in the paravertebral region after 5 months. Sutent (sutinitib) was initiated; however, due to hematological and gastrointestinal toxicities, the treatment was stopped after 2 weeks. He is currently receiving cabozantinib, and he is alive 14 months after the radical nephrectomy.
Pathological features
The tumors ranged in size from 3 to 10 cm (mean and median 7). The gross and histological features of the first seven cases have been previously described in detail [8]. Macroscopic examination of the radical nephrectomy specimen of case 8 revealed a 7 cm in greatest dimension solid mass with necrosis without renal vein invasion. Grossly, cases 9 and 10 were a well-circumscribed multicystic tumor of 3 cm in diameter (case 9) and a solid and cystic mass of 10 cm (case 10). The morphology of cases 8, 9 and 10 are illustrated separately for completeness.
Case 8
A discontinuous thick fibrous pseudocapsule was present. The tumor was predominantly composed of epithelioid cells with eosinophilic and focally clear cytoplasm and prominent nucleoli (G3 by ISUP/WHO 2016) mainly arranged in solid-alveolar architecture (Fig. 2). In some areas, smaller epithelioid clear cells were observed. Hemosiderin-laden histiocytes and extensive tumoral necrosis were noted.
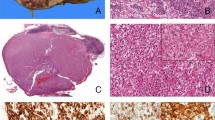
Histologic features of TFEB-amplified renal cell carcinomas. Low power view of case 8 shows a neoplasm with tumoral necrosis composed of epithelioid cells with eosinophilic (a) and clear cytoplasm and prominent nucleoli (b). The tumor of case 9 was composed of variably sized cysts (c) lined by a single layer of cuboidal cells with eosinophilic cytoplasm and small round nuclei (d). Two different architectures of case 10 were present, tubular-acinar (e) and solid areas made up of medium-sized polygonal cells with eosinophilic cytoplasm and prominent nucleoli (f)
Case 9
The tumor was well delineated by a fibrous pseudocapsule and characterized by a tubulocystic pattern with a thin eosinophilic fluid material filling the cystic spaces. The single layer of cuboidal cells lining the tubules and the cysts showed abundant granular eosinophilic cytoplasm with round nuclei and pinpoint nucleoli (G2 by ISUP/WHO 2016) (Fig. 2). Few macrophages bearing hemosiderin pigment were observed. Neither necrosis nor mitotic activity was found (< 1 per 10 High power field (HPF)).
Case 10
A thick fibrous pseudocapsule was present. The solid area of the neoplasm was mainly composed of medium-sized polygonal cells with eosinophilic and more rarely clear cytoplasm arranged in a alveolar and less frequently tubular-acinar or pseudopapillary (Fig. 2). The nuclei showed prominent nucleoli (G3 by ISUP/WHO 2016). Mitotic figures were occasionally encountered (0–1 per 10 HPF). Tumoral necrosis and hemorrhage were found.
Immunohistochemical features
The immunohistochemical results are tabulated in Table 2. As expected, all t(6;11) renal cell carcinomas were positive for cathepsin K, Melan-A and CK8-18 [9,10,11,12]. Staining for PAX8 and HMB45 was found in 7 of 8 tumors and in 6 of 8 tumors respectively (Fig. 3). Both TFEB-amplified renal cell carcinomas immunostained for PAX8, CK8-18 and cathepsin K, whereas just one tumor was positive for HMB45 and Melan-A. CD68 (PG-M1) was negative in all tumors.
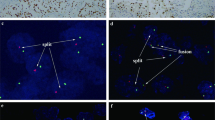
Immunophenotype of TFEB-amplified renal cell carcinomas. PAX8 was positive in the neoplastic cells of case 8 (a). The cells lined the cysts of case 9 were strongly positive for cathepsin K (high magnification in the insert) (b). Staining for Melan-A was diffusely present in the tumor cells of case 10 (c). Fluorescence in situ hybridization result of case 8 shows TFEB amplification (d). The green and red signals are split apart demonstrate the break of the TFEB gene (insert). TFEB gene amplification of case 10 (e) with identical level of amplification of VEGFA (f) and high VEGFA mRNA expression (g)
FISH results
All seven t(6;11) renal cell carcinomas and two metastasis demonstrated a high frequency of split TFEB fluorescent signals ranging from 61 to 94% (mean 74%, median 75%). In two tumors (cases 6 and 7), increased gene copy number was observed (3–5 fluorescent signals per neoplastic nuclei). Both tumors showed increased number of CEP6 (3–4 copies), whereas the remaining four tumors were disomic. The remaining three cases showed a high level of TFEB gene amplification (> 10 copies of fluorescent signals), one with TFEB rearrangement, the other two without evidence of rearrangement (Fig. 3). VEGFA was amplified in all three cases with TFEB amplification. In two of them (cases 8 and 10), the levels of amplification of VEGFA and TFEB were identical, whereas in case 9 the level of amplification of VEGFA was lower than the level of TFEB (Table 3).
RNAscope results
Overall, VEGFA mRNA expression was observed in 8 of 10 (80%) renal cell carcinomas with TFEB gene alteration (Table 3). Of these eight cases positive for VEGFA staining, three cases showed high-level TFEB amplification, one case showed TFEB rearrangement with increased TFEB gene copy number, whereas four showed TFEB gene rearrangement without increased copy number (Fig. 3). None of the papillary renal cell carcinomas, chromophobe renal cell carcinomas and oncocytomas demonstrated a positive staining for VEGFA, whereas two of three clear cell renal cell carcinomas showed a high VEGFA mRNA expression.
Discussion
An increasing number of manuscripts reported the presence of TFEB gene amplification in renal cell carcinoma [2,3,4,5,6]. As this tumor is defined by the occurrence of TFEB gene amplification what is considered amplified is of paramount importance. Gene amplification is established as an elevated extra copies of a gene without a proportional increase in other genes. Generally speaking, amplification of a gene may have diagnostic value (e.g., MDM2 amplification in well-differentiated liposarcoma) [13], prognostic value (e.g., MYC amplification in neuroblastoma) [14] or predictive value (e.g., HER-2 amplification in breast carcinoma) [15]. In renal cell carcinoma, TFEB gene amplification seems to be correlated with an aggressive behavior. The threshold proposed by Argani and coauthors is defined by the presence of an average of 10 or more copies per neoplastic nucleus [2]. Given the lack of a consensus to define TFEB amplification, Gupta et al. arbitrarily defined two levels of amplification, a low-level characterized by 5–10 copies and an high level with > 10 copies [4]. As the other studies used a cut-off of > 10 copies per nuclei, we decided to consider the latter as the threshold to use. Moreover, in previous analysis [2,3,4,5], no percentage of tumor cells harboring the amplification has been recorded, except in one recent study in which at least 10% of cells demonstrated the increasing fluorescent signals to consider the case amplified [6]. In the present series, the three tumors with TFEB amplification showed an increased gene copy number in virtually all neoplastic nuclei.
Another open and controversial issue is whether the increase of TFEB gene copy number is due to nonspecific whole DNA polyploidy vs. locus-specific TFEB amplification, in other words, whether it is a specific or nonspecific event. After corrections for centromeric alpha-satellite specific for chromosome 6, TFEB gene copy number was interpreted as true amplification rather than a nonspecific polyploidy in three cases (cases 8, 9 and 10). On the other hand, two cases (cases 6 and 7) showed a lower level of gene copy number (3–5 signals per tumor nuclei) with a similar increased number of CEP6. Hence, these were considered as chromosome 6 polysomy, a nonspecific event reflecting genomic instability.
With respect to the clinical behavior, we have previously reported two patients with t(6;11) renal cell carcinomas (cases 6 and 7) who developed metastasis characterized by increased TFEB gene copy number, necrosis and large tumor size [8]. As expected on the basis of pathologic features such as necrosis, dimension and amplification of TFEB gene, patient 8 recurred after 5 months and behaved aggressively.
Because of its novelty, the characteristics of TFEB-amplified renal cell carcinoma are not well understood; therefore, we undertook a comprehensive review of this tumor as illustrated in Table 4. Overall, 42 cases of TFEB-amplified renal cell carcinoma, including the three cases described herein, with or without TFEB rearrangement were found. The mean age of these patients was 63 years and the median 65 years (range from 23 to 83). There was a slight male predominance (24M, 18F). The tumors’ size ranged from 1.8 to 19.5 cm (mean and median 10). When follow-up was available, it ranged from 1 to 265 months (mean 79 and median 24). Based upon the review of the literature, the majority (26/41, 63%) of TFEB-amplified renal cell carcinomas are tumor stage pT3 or higher, which correlates with the aggressiveness. Moreover, most of the tumors (88%) showed an high ISUP/WHO 2016 nucleolar grade. Interestingly, as previously noted [4, 16] and reported here, TFEB amplification may occur in low-grade renal cell carcinoma. Histologically, the tumors with TFEB amplification were mainly characterized by a nested or papillary/psuedopapillary architecture made up of epithelioid cells with eosinophilic cytoplasm. None of the four t(6;11) renal cell carcinomas with concurrent TFEB gene amplification demonstrated the classical biphasic morphology with larger epithelioid cells and smaller cells clustered around eosinophilic spheres formed by basement membrane material. Nevertheless, the amplified – t(6;11) renal cell carcinoma (case 8) described in the present study showed two types of cells, large and small size. Immunohistochemically, labeling for HMB45, when reported present (6/27, 22%), is usually focal, whereas the positivity of cathepsin K (14/21, 67%) and Melan-A (26/33, 79%) ranged from patchy to diffuse. Among the four cases of t(6;11) renal cell carcinoma with concurrent TFEB gene amplification, all tumors expressed Melan-A, cathepsin K was present in 3 of 4, and half of them were labeled by HMB45. The expression of cathepsin K and melanocytic markers in TFEB-amplified renal cell carcinoma is worthy of note. A possible explanation is that, not only TFEB rearrangement but also increased TFEB gene copy number leads to intact TFEB protein overexpression, which correlates with aberrant melanocytic marker immunolabeling and cathepsin K expression as well.
In the current study, we have also assessed the occurrence of VEGFA amplification in renal cell carcinomas with TFEB gene alterations, either amplification or rearrangement. Increased VEGFA gene copy number (3–5 signals) was found in the two aggressive cases of t(6;11) renal cell carcinoma with a similar number of TFEB fluorescent signals. In the three TFEB-amplified renal cell carcinomas (> 10 TFEB signals), a concurrent VEGFA amplification was observed. The mRNA expression of VEGFA analyzed by RNAscope was concordant with VEGFA status in 7 out of 10 tumors (five VEGFA mRNA positive cases with VEGFA and TFEB gene copy number/amplification and two VEGFA mRNA negative cases with VEGFA and TFEB disomic status). In the remaining three cases, the level of VEGFA mRNA was higher than expected based on the level of VEGFA gene copy number suggesting the involvement of an alternative mechanism leading the upregulation of mRNA expression.
The last fascinating aspect regards the treatment. At present, there are few studies concerning the target therapy (mammalian target of rapamycin (mTOR) inhibitor and anti-angiogenic including anti-VEGF receptor and ligand) in MiT family translocation renal cell carcinoma [17, 18]. It is important to note that all the tumors of the patients reported by Malouf et al. and Choueiri et al. were Xp11 renal cell carcinoma and data regarding the treatment of aggressive t(6;11) renal cell carcinoma are lacking. Recently, Gupta and colleagues described the possible usefulness of VEGFR target therapy in four renal cell carcinomas with TFEB/VEGFA coamplification [4]. This finding is interesting because the t(6;11) renal cell carcinoma with aggressive behavior reported in the present study were characterized by increased TFEB/VEGFA gene copy number suggesting that VEGFA may be a potential therapeutic target in this subset of tumors.
In summary, we have described a series of 10 renal cell carcinomas with TFEB gene molecular alterations and immunoexpression of cathepsin K including seven cases of unamplified conventional t(6;11) renal cell carcinomas, two of which had increased TFEB/VEGFA gene copy number, one amplified t(6;11) renal cell carcinoma with concurrent VEGFA amplification and two TFEB/VEGFA-amplified renal cell carcinomas without TFEB gene rearrangement. On the basis of those findings, one could suggest that all the renal tumors showing morphological characteristics suggesting t(6;11) renal cell carcinoma and all unclassified renal cell carcinomas, either high grade or low grade, should immunohistochemically be evaluated for cathepsin K and/or Melan-A and if one of them is positive, tested for TFEB gene alteration and VEGFA gene amplification. Finally, we suggest VEGFA as a potential therapeutic target in aggressive renal cell carcinoma with TFEB gene alterations.
References
Argani P. MiT family translocation renal cell carcinoma. Semin Diagn Pathol. 2015;32:103–13.
Argani P, Reuter VE, Zhang L, Sung YS, Ning Y, Epstein JI, et al. TFEB-amplified renal cell carcinomas: an aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am J Surg Pathol. 2016;40:1484–95.
Williamson SR, Grignon DJ, Cheng L, Favazza L, Gondim DD, Carskadon S, et al. Renal cell carcinoma with chromosome 6p amplification including the TFEB gene: a novel mechanism of tumor pathogenesis? Am J Surg Pathol. 2017;41:287–98.
Gupta S, Johnson SH, Vasmatzis G, Porath B, Rustin JG, Rao P, et al. TFEB-VEGFA (6p21.1) co-amplified renal cell carcinoma: a distinct entity with potential implications for clinical management. Mod Pathol. 2017;30:998–1012.
Skala SL, Xiao H, Udager AM, Dhanasekaran SM, Shukla S, Zhang Y, et al. Detection of 6 TFEB-amplified renal cell carcinomas and 25 renal cell carcinomas with MITF translocations: systematic morphologic analysis of 85 cases evaluated by clinical TFE3 and TFEB FISH assays. Mod Pathol. 2018;31:179–97.
Mendel L, Ambrosetti D, Bodokh Y, Ngo-Mai M, Durand M, Simbsler-Michel C, et al. Comprehensive study of three novel cases of TFEB-amplified renal cell carcinoma and review of the literature: evidence for a specific entity with poor outcome. Genes Chromosomes Cancer. 2018;57:99–113.
Peckova K, Vanecek T, Martinek P, Spagnolo D, Kuroda N, Brunelli M, et al. Aggressive and nonaggressive translocation t(6;11) renal cell carcinoma: comparative study of 6 cases and review of the literature. Ann Diagn Pathol. 2014;18:351–7.
Calio A, Brunelli M, Segala D, Pedron S, Tardanico R, Remo A, et al. t(6;11) renal cell carcinoma: a study of seven cases including two with aggressive behavior, and utility of CD68 (PG-M1) in the differential diagnosis with pure epithelioid PEComa/epithelioid angiomyolipoma. Mod Pathol. 2018;31:474–87.
Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22:1016–22.
Argani P, Lae M, Hutchinson B, Reuter VE, Collins MH, Perentesis J, et al. Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am J Surg Pathol. 2005;29:230–40.
Argani P, Yonescu R, Morsberger L, Morris K, Netto GJ, Smith N, et al. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am J Surg Pathol. 2012;36:1516–26.
Smith NE, Illei PB, Allaf M, Gonzalez N, Morris K, Hicks J, et al. t(6;11) renal cell carcinoma (RCC): expanded immunohistochemical profile emphasizing novel RCC markers and report of 10 new genetically confirmed cases. Am J Surg Pathol. 2014;38:604–14.
Zhang H, Erickson-Johnson M, Wang X, Oliveira JL, Nascimento AG, Sim FH, et al. Molecular testing for lipomatous tumors: critical analysis and test recommendations based on the analysis of 405 extremity-based tumors. Am J Surg Pathol. 2010;34:1304–11.
Canete A, Gerrard M, Rubie H, Castel V, Di Cataldo A, Munzer C, et al. Poor survival for infants with MYCN-amplified metastatic neuroblastoma despite intensified treatment: the International Society of Paediatric Oncology European Neuroblastoma Experience. J Clin Oncol. 2009;27:1014–9.
Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92.
Durinck S, Stawiski EW, Pavia-Jimenez A, Modrusan Z, Kapur P, Jaiswal BS, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015;47:13–21.
Choueiri TK, Lim ZD, Hirsch MS, Tamboli P, Jonasch E, McDermott DF, et al. Vascular endothelial growth factor-targeted therapy for the treatment of adult metastatic Xp11.2 translocation renal cell carcinoma. Cancer . 2010;116:5219–25.
Malouf GG, Camparo P, Oudard S, Schleiermacher G, Theodore C, Rustine A, et al. Targeted agents in metastatic Xp11 translocation/TFE3 gene fusion renal cell carcinoma (RCC): a report from the Juvenile RCC Network. Ann Oncol. 2010;21:1834–8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Caliò, A., Brunelli, M., Segala, D. et al. VEGFA amplification/increased gene copy number and VEGFA mRNA expression in renal cell carcinoma with TFEB gene alterations. Mod Pathol 32, 258–268 (2019). https://doi.org/10.1038/s41379-018-0128-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0128-1
This article is cited by
-
mTOR eosinophilic renal cell carcinoma: a distinctive tumor characterized by mTOR mutation, loss of chromosome 1, cathepsin-K expression, and response to target therapy
Virchows Archiv (2023)
-
Circ-AFAP1 promote clear cell renal cell carcinoma growth and angiogenesis by the Circ-AFAP1/miR-374b-3p/VEGFA signaling axis
Cell Death Discovery (2022)
-
TSC loss is a clonal event in eosinophilic solid and cystic renal cell carcinoma: a multiregional tumor sampling study
Modern Pathology (2022)
-
TFE3 and TFEB-rearranged renal cell carcinomas: an immunohistochemical panel to differentiate from common renal cell neoplasms
Virchows Archiv (2022)
-
Diagnostic utility of one-stop fusion gene panel to detect TFE3/TFEB gene rearrangement and amplification in renal cell carcinomas
Modern Pathology (2021)