Abstract
Estrogen receptor-α (ER-α), encoded by ESR1, is detected by immunohistochemistry in approximately 70% of invasive breast cancers and serves as a strong predictive biomarker. ESR1-activating mutations in the ligand-binding domain have been reported in up to 35–40% of ER-positive metastatic breast cancers and are associated with endocrine therapy resistance and disease progression. At present, it is unclear whether ESR1 mutations alter the immunohistochemical detection of ER performed in routine clinical practice. In this study, ESR1 mutations in breast cancer were identified utilizing Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), a Food and Drug Administration-approved hybridization capture-based next-generation sequencing assay. Five hundred and eighty-six breast cancers from patients with locally advanced or metastatic disease were analyzed using MSK-IMPACT in the study period. ESR1 somatic alterations were identified in 67 breast cancer samples from 66 patients. Immunohistochemical analysis of ER, progesterone receptor, and human epidermal growth factor receptor 2 was performed on the primary and treated breast cancers from these patients at the time of diagnosis. Twenty unique ESR1 mutations were identified involving the ligand-binding domain, all in breast cancer samples from patients previously treated with endocrine therapy. The most frequent mutations were D538G (n = 22), Y537S (n = 7), and E380Q (n = 7). All breast cancer samples with an ESR1 mutation were ER-positive by immunohistochemistry. Review of the ER immunohistochemistry in the paired untreated primary tumor and treated tumor from 34 patients showed no detectable change in the ER-positive immunohistochemical status (median percentage of invasive tumor cells with nuclear staining: untreated primary tumor 90%, treated tumor 95%). We conclude that ESR1 mutations do not appreciably diminish ER-positive staining by immunohistochemistry. In addition to standard biomarker testing by immunohistochemistry, the assessment of ESR1 mutations by molecular testing can help guide the clinical management of patients with ER-positive breast cancer in the setting of endocrine resistance and progression of disease.
Similar content being viewed by others
Introduction
Estrogen receptor-α (ER-α), encoded by ESR1, is a ligand-activated transcription factor that dimerizes and localizes to the nucleus. Approximately 70% of invasive breast cancers express ER-α. The detection of ER by immunohistochemistry determines the use of endocrine therapy, and studies have shown the ER status to be a weak prognostic but strong predictive biomarker in patients with ER-positive tumors [1,2,3].
Antiestrogen therapy has been an effective treatment strategy for women with ER-positive breast cancer [4, 5] and includes estrogen deprivation therapy (aromatase inhibitors, gonadotropin-releasing hormone agonists) and direct inhibitors of ER (selective ER modulators and selective ER degraders) [6, 7]. Despite the benefits of these drugs, a large proportion of patients with ER-positive breast cancer will develop resistance after prolonged exposure [8,9,10,11]. Somatic mutations involving the ligand-binding domain of the ESR1 gene are reported in ER-positive breast cancer after prolonged estrogen deprivation therapy [12,13,14]. Activating ESR1 mutations occur in up to 35–40% of hormone-resistant ER-positive breast cancer, including studies assessing tissue samples and circulating tumor DNA, and confer constitutive ligand-independent activation of ER transcription and ER-α expression, mediating antiestrogen resistance [12,13,14,15,16,17,18,19]. The most common ESR1 mutations are D538G and Y537S, which are reported as being associated with a worse overall prognosis [16]. These mutations rarely occur in untreated primary tumors, but were detected in nearly 30% of ER-positive metastatic breast cancer, previously treated with aromatase inhibitors [15, 16]. These findings indicate that certain mutations in the ligand-binding domain of ESR1 result in acquired resistance to endocrine therapy and an alternate treatment may be warranted.
The assessment of ER status is routinely performed by immunohistochemistry on primary breast cancers, breast cancer recurrences, and metastases. In the era of molecular testing, ESR1 somatic alterations can now be detected by hot-spot mutation panels or comprehensive next-generation sequencing assays. At this time, it is unknown whether acquired ESR1 mutations affect the detection of ER by conventional immunohistochemistry. In this study, we assessed ER expression by immunohistochemistry in breast cancer samples with ESR1 mutations detected by next-generation sequencing.
Materials and methods
Case selection and standard receptor status testing
After obtaining institutional review board approval, the database of the Molecular Diagnostics service was searched for breast cancers with ESR1 mutations detected by the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay [20, 21]. Immunohistochemical testing for ER, progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) was performed and reported at the time of diagnosis on primary, recurrent, and metastatic tumors according to the ASCO/CAP guideline recommendations [22, 23] using a Food and Drug Administration (FDA)-cleared/approved method (Table 1). The percentage of nuclear staining, intensity of nuclear staining, and antibody utilized for ER and PR detection was recorded for all available cases. Few patients had initial ER and PR testing on the primary breast cancer performed by either flow cytometry or ligand-binding assays. Reflex HER2 fluorescence in situ hybridization (FISH) testing was performed on cases with an equivocal HER2 immunohistochemical result (HER2 IQFISH pharmDx™, Dako, Carpinteria, CA, USA; PathVysion HER2 DNA Probe Kit, Vysis, Downers Grove, IL, USA). Hormone receptor analysis was performed on the same specimen as that submitted for next-generation sequencing testing when sufficient material was available. All available diagnostic slides were reviewed by three pathologists (DSR, FK, HYW) to confirm the ER, PR, and HER2 immunohistochemistry results.
Information about tumor size, grade, and histologic subtype of the primary breast cancer was extracted from the pathology reports. Clinical information, including treatment, for all patients was retrieved from the electronic medical records.
The MSK-IMPACT assay
Details of the MSK-IMPACT assay have been previously published [20, 21]. MSK-IMPACT is a comprehensive molecular profiling assay that involves hybridization capture and deep sequencing of all exons and selected introns of up to 410 oncogenes and tumor suppressor genes during this study period, allowing the detection of point mutations, small and large insertions or deletions, as well as rearrangements. In addition to capturing all coding regions of the genes, the assay also captures over 1000 intergenic and intronic single-nucleotide polymorphisms (tiling probes), interspersed homogenously across the genome, aiding the accurate assessment of genome-wide copy number. In total, the probes target approximately 1.2 Mb of the human genome.
Tested samples were confirmed to contain at least 10% tumor content (i.e., percentage of tumor cells relative to benign epithelial cells, stromal cells, and inflammatory cells) by pathologist review of hematoxylin and eosinstained sections from the same tissue block submitted for DNA extraction. Genomic DNA was extracted from formalin-fixed, paraffin-embedded tumor tissue after manual macrodissection. DNA from matching peripheral blood (EDTA tube) for each patient was also extracted and used as a normal control. Sequence libraries were prepared (Kapa Biosystems, Wilmington, MA, USA) through a series of enzymatic steps including shearing of double-stranded DNA, end repair, A-base addition, ligation of barcoded sequence adaptors, and low-cycle PCR amplification. Multiple barcoded sequence libraries were pooled and captured using our custom-designed biotinylated probes (Roche NimbleGen, Madison, WI, USA). Captured DNA fragments were sequenced on an Illumina HiSeq2500 (San Diego, CA, USA) as 100 bp paired-end reads, which were then subject to the bioinformatics analysis pipeline as described by Cheng et al. [20].
Results
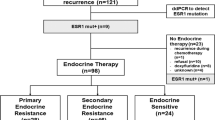
A total of 586 breast cancers from patients with locally advanced or metastatic disease were sequenced during the study time period (1/2014–5/2015). ESR1 somatic alterations were identified in 67 breast cancer samples from 66 patients (one patient had two metastatic tumors tested). Figure 1 summarizes the breakdown of samples with ESR1 alterations. The majority (n = 56) of the samples in this data set were metastatic tumors previously treated with endocrine therapy. One metastatic tumor was from a patient with no documented history of endocrine therapy. In 10 patients, ESR1 somatic alterations were detected in the primary breast cancer samples, including five untreated primary tumors and five primary tumors status post endocrine therapy for previously diagnosed breast cancer (three patients with a history of contralateral breast cancer and two patients presenting with advanced stage disease, treated with neoadjuvant endocrine and chemotherapy). The metastatic tumors for these 10 patients were not suitable or not available for MSK-IMPACT testing.
Table 2 summarizes the clinicopathologic characteristics of the patients. Median age at primary breast cancer diagnosis was 47 years (range 24–75 years). The primary breast cancer showed ductal (n = 53) and lobular (n = 12) histology. One patient had a history of ductal carcinoma in situ (DCIS) only and developed distant metastasis 11 years later. Median primary tumor size was 2.8 cm among the 52 patients with available tumor size information and 18 primary tumors were multifocal. The primary tumor grade was well differentiated (7%), moderately differentiated (30%), moderately to poorly differentiated (9%), and poorly differentiated (54%) in the 43 patients with available information. The receptor status for the primary tumors with available information are: 97% (59/61) ER-positive, 88% (51/58) PR-positive, and 13% (7/53) HER2-positive. The two primary tumors with a reported ER-negative status were PR-positive.
The breakdown of the endocrine therapy used in the 60 patients who had a tumor analyzed status post treatment is listed in Table 2. All patients had prior exposure to aromatase inhibitors, with 67% of patients receiving both aromatase inhibitors and tamoxifen during the course of their treatment. Of note, 39 patients developed metastatic or recurrent disease while on endocrine therapy. The median interval from primary breast cancer diagnosis to metastasis was 66 months (range 1–263). Seven patients presented with advanced stage disease.
Hormone receptor results for the tumor status post endocrine therapy by immunohistochemistry was available on the same specimen sequenced by MSK-IMPACT in all but eight cases; the receptor results for these eight cases was obtained from an alternate tumor status post treatment. The receptor results for the tumors from patients in this cohort status post endocrine therapy are: 100% (65/65) ER-positive, 76% (47/62) PR-positive, and 5% (3/63) HER2-positive (Table 2). The median and mean percentage of invasive tumor cells with nuclear staining was 95 and 87%, respectively, for ER and 20 and 43%, respectively, for PR. Of note, not included in this category are one primary tumor and one metastatic tumor without prior endocrine therapy and no available results on a subsequent tumor status post treatment.
ESR1 mutations in this cohort included 60 single-nucleotide variants (SNVs) and 11 copy number alterations (10 whole-gene gains/amplifications, one whole-gene deletion) for a total of 71 ESR1 somatic alterations. All tumor samples showed adequate coverage, with an average depth of 623× (range 142–1971). Of the 60 ESR1 SNVs, 20 unique SNVs were identified, all involving the ligand-binding domain (Fig. 2 and Supplemental Table 1). The most frequent SNVs were D538G (n = 22), Y537S (n = 7), and E380Q (n = 7). A total of 40 SNVs affected amino acids L536 to D538 (known activating mutations) and all were identified in post endocrine therapy breast cancer samples. The ESR1 mutations identified in the five primary tumors and one metastatic tumor with no prior endocrine therapy include: S432L, K520fs, three whole-gene amplifications, and one whole-gene deletion.
The assay also uncovered concurrent mutations in 28 cancer genes, most common including PIK3CA, GATA3, CDH1, and TP53 (Fig. 3).
Semi-quantitative review of the ER and PR immunohistochemistry was available in the paired untreated primary tumor (Tumor 1) and treated tumor (metastasis, primary tumor status post endocrine therapy for contralateral breast cancer, or neoadjuvant therapy) (Tumor 2) for 34 patients (Supplemental Table 2). The ER and PR immunohistochemical results and the antibodies used (if available) for these paired tumors are listed in Supplemental Table 2. The amount of material available for analysis in the metastatic tumor was taken into account, since some tumors in the metastatic sample were scant. We observed no detectable change of the ER status detected by immunohistochemistry in this subset. For ER, the median and mean percentage of invasive tumor cells with nuclear staining was 90 and 86%, respectively, for Tumor 1, and 95 and 90%, respectively, for Tumor 2. For PR, the median and mean percentage of invasive tumor cells with nuclear staining was 35 and 42%, respectively, for Tumor 1, and 70 and 55%, respectively, for Tumor 2. Three metastatic tumors showed loss of PR positivity compared to the primary breast cancer and one metastasis showed substantially decreased PR positivity by immunohistochemistry. The ESR1 mutations for these four cases are listed in Table 3. The immunohistochemistry results for a representative case are illustrated in Fig. 4.
Representative case with an ESR1 p.Y537S (c.1610A>C) mutation. a Hematoxylin and eosinstained (H&E) section of metastatic carcinoma involving the epidural space (×400). b Estrogen receptor (ER) immunohistochemical-stained slide demonstrated 60% nuclear staining with strong intensity (×400). c Progesterone receptor (PR) immunohistochemical-stained slide demonstrated 90% nuclear staining with strong intensity (×400)
Discussion
Somatic mutations in the ligand-binding domain of the ESR1 gene are found in ER-positive breast cancer after prolonged endocrine therapy, leading to acquired resistance to aromatase inhibitors and disease progression [12,13,14,15,16]. In this study, we assessed ER status by immunohistochemistry in breast cancer samples with ESR1 mutations detected by next-generation sequencing. This study demonstrated that ER remains positive by immunohistochemistry in breast cancer with acquired ESR1 mutations. Although assessing ER/PR status by immunohistochemistry assists in confirming the diagnosis of recurrent/metastatic breast cancer, a positive result no longer predicts benefit from all endocrine therapy if an ESR1 mutation is present. Persistent positive staining in metastatic disease with a documented ESR1 mutation suggests treatment with an alternate endocrine therapy, such as a selective ER degrader, is indicated.
Functional studies for the majority of the ESR1 SNVs detected in this study have been published by Toy et al. [24]. The most common mutations within the ligand-binding domain in this cohort were D538G, Y537S, and E380Q, which together accounted for 51% (36/71) of all ESR1 somatic alterations. These mutations have been shown to favor the receptor’s agonist confirmation by promoting a configuration of Helix 12 despite the absence of ligand [12,13,14, 25]. The functional significance of these ligand-binding domain mutations has been supported by in vitro models in which higher levels of ER-regulated genes are expressed in the absence of ligand [12]. Additionally, in vivo models have shown many ESR1 ligand-binding domain mutations that render resistance to estrogen deprivation via constitutive activity however only select activating mutations such as Y537S caused a magnitude of change associated with fulvestrant resistance [24]. Correspondingly, tumors driven by Y537S, but not D538G, E380Q, or S463P, were less effectively inhibited by fulvestrant than more potent and bioavailable antagonists, including AZD9496 [24].
ESR1 mutations are rarely detected in the primary tumor (<1% in The Cancer Genome Atlas data set) [26], as it is believed that the mutations develop as an acquired endocrine resistance mechanism. In this study, the ESR1 alterations were identified in five primary tumors and one metastatic tumor with no prior endocrine therapy, including S432L, K520fs, three whole-gene amplifications, and one whole-gene deletion. The S432L mutant has been shown to have little evidence of constitutive activity in the absence of estradiol [24] however the significance of the other alterations is unclear at this time.
The ER and PR status serves as valuable predictive biomarker in breast cancer and immunohistochemical testing guides adjuvant endocrine therapy. FDA-cleared ER antibodies for breast cancer testing by immunohistochemistry include clones 1D5, 6F11, SP1, and ER.2.123 + 1D5 (cocktail) [22]. Standardization of the reagents and assay improves reproducibility and reduces inaccurate test results. Each of these antibodies has been clinically validated, demonstrating good correlation with patient outcomes and benefit from adjuvant endocrine therapy [22]. The various ER antibody clones recognize different epitopes however none of the FDA-cleared ER antibodies binds an epitope within the ligand-binding domain. Clones 6F11 and ER.2.123 + 1D5 recognize the N-terminus (6F11 and ER.2.123: amino acids 15–23; 1D5: amino acids 127–130), whereas SP1 recognizes the C-terminus (amino acids 578–595) [27, 28]. ESR1 mutations affecting the ligand-binding domain, therefore, do not appear to alter the epitope structure or antibody binding to the receptor. This is consistent with our findings that there was no observed change in ER status assessed by immunohistochemistry in breast cancers with acquired ESR1 mutations in the ligand-binding domain.
In this study, the ER staining between Tumor 1 (untreated) and Tumor 2 (treated) in the 34 paired tumors was similar (median percentage of invasive tumor cells with nuclear staining: Tumor 1 90%; Tumor 2 95%), whereas the median percentage for PR staining was 35% for Tumor 1 and 70% for Tumor 2. The overall higher expression of PR by immunohistochemistry in samples with acquired ESR1 mutations is consistent with data recently reported in a metastatic breast cancer patient from a rapid autopsy cohort [29, 30]. The enhanced expression of PR was also observed in the CRISPR knock-in ESR1 Y537S cell line [24].
In summary, our results demonstrate that ER immunohistochemistry is not predictive of ESR1 mutation status as all tumors in this study with an ESR1-activating mutation were ER-positive. Formalin-fixed, paraffin-embedded tumor samples are often used for molecular testing however ESR1 mutation detection in cell-free DNA by droplet digital PCR analysis also shows promising data [16, 31]. In addition to standard biomarker testing by immunohistochemistry, the assessment of ESR1 mutations by molecular testing can help guide the clinical management of patients with ER-positive breast cancer in the setting of endocrine resistance and progression of disease.
References
Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–81.
Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–68.
Allred DC. Issues and updates: evaluating estrogen receptor-alpha, progesterone receptor, and HER2 in breast cancer. Mod Pathol. 2010;23:S52–9.
EBCTCG. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717.
Mouridsen H, Gershanovich M, Sun Y, et al. Phase III study of letrozole versus tamoxifen as first-line therapy of advanced breast cancer in postmenopausal women: analysis of survival and update of efficacy from the International Letrozole Breast Cancer Group. J Clin Oncol. 2003;21:2101–9.
Ariazi EA, Ariazi JL, Cordera F, Jordan VC. Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem. 2006;6:181–202.
Patel HK, Bihani T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol Ther. 2018;186:1–24.
Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–43.
Strasser-Weippl K, Goss PE. Advances in adjuvant hormonal therapy for postmenopausal women. J Clin Oncol. 2005;23:1751–9.
Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53.
Peng J, Sengupta S, Jordan VC. Potential of selective estrogen receptor modulators as treatments and preventives of breast cancer. Anticancer Agents Med Chem. 2009;9:481–99.
Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–45.
Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–51.
Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20:1757–67.
Fribbens C, O’Leary B, Kilburn L, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016;34:2961–8.
Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016;2:1310–5.
Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-alpha: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73:6856–64.
Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579.
Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7:313ra182.
Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–64.
Zehir A, Benayed R, Shah RH, et al. Erratum: mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:1004.
Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95.
Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013.
Toy W, Weir H, Razavi P, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7:277–87.
Fanning SW, Mayne CG, Dharmarajan V, et al. Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. Elife. 2016;5:e12792.
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Kornaga EN, Klimowicz AC, Guggisberg N, et al. A systematic comparison of three commercial estrogen receptor assays in a single clinical outcome breast cancer cohort. Mod Pathol. 2016;29:799–809.
Brock JE, Hornick JL, Richardson AL, Dillon DA, Lester SC. A comparison of estrogen receptor SP1 and 1D5 monoclonal antibodies in routine clinical use reveals similar staining results. Am J Clin Pathol. 2009;132:396–401.
Wu JM, Fackler MJ, Halushka MK, et al. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–46.
Avigdor BE, Cimino-Mathews A, DeMarzo AM, et al. Mutational profiles of breast cancer metastases from a rapid autopsy series reveal multiple evolutionary trajectories. JCI Insight. 2017;2(24):e96896
Wang P, Bahreini A, Gyanchandani R, et al. Sensitive detection of mono- and polyclonal ESR1 Mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin Cancer Res. 2016;22:1130–7.
Acknowledgements
This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748 and supported by the Marie-Josée and Henry R Kravis Center for Molecular Oncology. SC has grant support by NIH R01CA204999.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have read and approved the manuscript and have contributed sufficiently to the project to be included as authors. The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ross, D.S., Zehir, A., Brogi, E. et al. Immunohistochemical analysis of estrogen receptor in breast cancer with ESR1 mutations detected by hybrid capture-based next-generation sequencing. Mod Pathol 32, 81–87 (2019). https://doi.org/10.1038/s41379-018-0116-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0116-5
This article is cited by
-
MEN1 mutations mediate clinical resistance to menin inhibition
Nature (2023)
-
Heterogeneity of estrogen receptor based on 18F-FES PET imaging in breast cancer patients
Clinical and Translational Imaging (2021)