Abstract
Metaplastic breast carcinomas comprise a histologically heterogenous group of tumors. Although most are triple (estrogen/progesterone receptor, HER2) negative, these rare tumors are clinicopathologically distinct from other triple negative carcinomas and may be aggressive with worse chemotherapy responses. On the other hand, metaplastic carcinomas are histologically diverse, which is reflected in gene expression differences among subtypes. Whether metaplastic carcinomas are genetically distinct from other triple negative cancers and whether genetic differences underlie histologic subtypes remains poorly understood. We sequenced 408 cancer-related genes in 28 metaplastic carcinomas, including chondroid matrix-producing carcinomas (n = 10), spindle cell carcinomas (n = 5), and carcinomas with squamous (n = 5), mixed spindle/squamous (n = 5), and mixed metaplastic (n = 3) differentiation. Metaplastic carcinomas were highly enriched for PIK3CA/PIK3R1 (61%) and Ras-Map kinase (25%) pathway aberrations compared to other triple negative carcinomas (TCGA dataset 14%, p < 0.001 and 7%, p = 0.005, respectively) and harbored a high frequency of TP53 (64%) and TERT promoter (25%) mutations, but this varied among subtypes. Chondroid-matrix producing carcinomas lacked PI-3 kinase and Ras-Map kinase aberrations and TERT promoter mutations, compared to 100%, 39%, and 39% of non-matrix-producing tumors, respectively. TERT promoter mutations were enriched (47%) in spindle cell carcinomas and tumors with squamous or spindle/squamous differentiation. Spindle cell carcinomas lacked TP53 mutations, in contrast to other subtypes (78%, p = 0.003). Separate analysis of paired ductal carcinoma in situ and metaplastic carcinoma revealed shared clonality in all cases (n = 8). Activating PI-3 kinase and Ras pathway mutations were early events, and inactivating mutations in tumor suppressors including RB1, CDKN2A, and TP53 were associated with invasion in individual cases. Metaplastic components of two tumors showed genetic progression from separately sequenced paired invasive ductal carcinoma. The findings suggest that metaplastic carcinomas are genetically distinct from other triple negative breast cancers and highlight genetic heterogeneity that broadly correlates with histologic subtype. Heterologous elements progress from associated ductal carcinoma.
Similar content being viewed by others
Introduction
Metaplastic breast carcinomas are a rare and histologically diverse group of breast cancers, estimated to comprise 0.2–5% of invasive carcinomas in the breast [1]. As a group, these tumors are defined histologically by differentiation of neoplastic epithelium into squamous or mesenchymal-like elements, including spindle cell and heterologous chondroid, osseous, or rhabdomyoid differentiation, among less common types [1,2,3,4,5]. A subgroup of cartilaginous and/or osseous matrix-producing carcinomas lacking a spindle cell component has been recognized as histologically distinct from other tumors with heterologous differentiation [5, 6]. The metaplastic component may arise with or without an associated in situ or invasive ductal component [7]. This heterogeneity is further highlighted by recognition of low-grade (fibromatosis-like and adenosquamous carcinoma) and more aggressive high-grade variants, which comprise the majority of tumors [1]. Most metaplastic breast carcinomas lack expression of estrogen receptor (ER), progesterone receptor (PR), and HER2 (triple negative phenotype), but are considered to have more aggressive behavior, worse outcomes, and poorer response to chemotherapy than other triple negative invasive ductal carcinomas of no special type [7,8,9,10,11,12]. Clinicopathologic features of metaplastic breast carcinomas are also distinct from other triple negative breast cancers, including presentation with larger tumor size, less frequent axillary lymph node involvement, and more common local recurrence/distant metastatic spread [7,8,9, 12, 13]. Histologic subtype has been identified in some studies as an independent prognostic feature [3,4,5, 7, 14]. Nonetheless, management and treatment of aggressive metaplastic breast carcinoma variants is typically similar between histologic subtypes and similar to other high grade triple negative carcinomas [2, 15].
The molecular features of metaplastic breast carcinomas are poorly defined, and the underlying basis for histologic heterogeneity remains uncertain. As a group, the majority of metaplastic breast carcinomas demonstrate basal-like or claudin-low gene expression profiles [16,17,18,19]. However, these tumors are transcriptomically heterogeneous within and between histologic subtypes. For instance, in contrast to other histologic subtypes, spindle cell carcinomas are uniformly claudin-low by expression profiling with lower expression of genes involved in epithelial-to-mesenchymal transition, whereas tumors with chondroid metaplasia are more homogeneously mesenchymal stem-like by triple negative breast cancer subtyping [17, 20]. Little is known regarding the genetics of metaplastic breast carcinomas. Some studies have demonstrated a high frequency of phosphoinositide (PI)-3 kinase pathway aberrations including frequent PIK3CA mutations, as well as TP53 mutations, in a background of more heterogeneous lower frequency aberrations, such as CDKN2A loss, EGFR amplification, and Wnt pathway aberrations [18, 20,21,22,23,24,25,26]. Whether histologic subtypes are associated with distinct underlying genomic features and oncogenic driver aberrations compared to other subtypes remains largely unexplored but may help explain differences in behavior and outcome [3,4,5, 7, 14, 24]. A better understanding of pathogenesis and genetics may also lead to better treatment approaches for these aggressive tumors.
Although early studies demonstrated shared clonality of different histologic components in these tumors [27,28,29,30,31], the degree of genetic relatedness of different heterologous elements to one another and to associated invasive or in situ ductal carcinoma remains poorly characterized. In fact, the repertoire of somatic mutations of ductal carcinoma in situ (DCIS) associated with metaplastic carcinomas has not been investigated and could shed light on tumor histogenesis.
In this study, we used capture-based next-generation sequencing of 408 cancer-related genes in order to comprehensively characterize the genomics of metaplastic breast carcinomas and to compare the genetics of various histologic subtypes with one another and with publically available data of other triple negative breast cancers. Metaplastic components in a subset of tumors were also compared with associated in situ and invasive ductal carcinoma in order to determine the genetic relationship between these components. The findings shed light on our understanding of the biology and progression of metaplastic breast carcinomas and highlight the underlying genetic heterogeneity of these rare aggressive tumors.
Materials and methods
Study population
This study was approved by the institutional review board of the University of California San Francisco. Twenty-eight metaplastic breast carcinomas, spanning years 2003–2017, were identified in the Pathology department archives. Low grade fibromatosis-like and adenosquamous carcinomas were excluded. All cases were reviewed by G.K. and Y-Y.C to confirm the diagnosis and lines of differentiation. All specimens were fixed in 10% neutral buffered formalin and embedded in paraffin. Clinical information was obtained from the electronic medical record of UCSF or referring facilities when applicable.
Capture-based next generation DNA sequencing
Matched normal and tumor tissue was selected from 23 metaplastic carcinomas for capture-based next-generation DNA sequencing. For an additional five cases (Sp2, Sp3, Sq3, Sq5, and SS2), only tumor tissue was selected for sequencing. For eight tumors, DNA from associated DCIS was extracted and analyzed separately. For three tumors (including two for which DCIS was analyzed separately), DNA was extracted and analyzed from two separate areas with differing lines of differentiation. Sequencing libraries were prepared from genomic DNA extracted from tumor and normal formalin-fixed paraffin embedded tissue. Target enrichment was performed by hybrid capture using a custom oligonucleotide library. Sequencing was performed at the UCSF Clinical Cancer Genomics Laboratory using an assay that targets the coding regions of 408 cancer-related genes, select introns from 42 genes, and TERT promoter with a total sequencing footprint of 2.8 Mb (Supplemental Table S1) [32, 33]. Sequencing was performed on an Illumina HiSeq 2500. Duplicate sequencing reads were removed computationally to allow for accurate allele frequency determination and copy number calling. The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37), using the following software packages: BWA: 0.7.10-r789, Samtools: 1.1 (using htslib 1.1), Picard tools: 1.97 (1504), GATK: 2014.4-3.3.0-0-ga3711, CNVkit: 0.3.3, Pindel: 0.2.5a7, SATK: 2013.1-10- gd6fa6c3, Annovar: v2015Mar22, Freebayes: 0.9.20 and Delly: 0.5.9 [34,35,36,37,38,39,40,41,42,43,44]. Only insertions/deletions (indels) up to 100 base pairs in length were included in the mutational analysis. Somatic single nucleotide variants and insertions/deletions were visualized and verified using Integrated Genome Viewer. Genome-wide copy number analysis based on on-target and off-target reads was performed by CNVkit and Nexus Copy Number (Biodiscovery, Hawthorne, CA) [38]. Tumors treated with neoadjuvant chemotherapy prior to sequencing were excluded from copy number analysis.
Results of DNA sequencing were compared to publicly available sequencing data from breast carcinomas in The Cancer Genome Atlas (TCGA) sorted for triple negative status [45, 46]. Statistical comparisons between metaplastic carcinomas and TCGA breast cancers and between metaplastic carcinoma histologic subtypes were performed using Fisher exact, chi-squared, and student’s t-tests using a level of significance of p < 0.05.
Immunohistochemistry
Immunohistochemistry for ER, PR, and HER2 was performed on whole tissue sections using CC1 (Roche, Ventana Medical Systems) for antigen retrieval and clones SP1, 1E2, and 4B5, respectively (all undiluted; Roche, Ventana Medical Systems, Tucson, AZ). Positive staining was defined according to ASCO/CAP guidelines.
Results
Clinicopathologic features of metaplastic carcinomas
Clinicopathologic features of metaplastic carcinomas included in this study are shown in Table 1. Patient ages ranged from 25 to 85 years (mean 61, median 64 years). Cases included five pure spindle cell carcinomas (Sp1-5), five carcinomas with squamous differentiation (Sq1-5), five carcinomas with mixed spindle and squamous differentiation (SS1-5), ten matrix-producing carcinomas with chondroid differentiation, one of which also had a minute focus (<5%) of associated bone (C1-10), and three carcinomas with mixed heterologous differentiation (M1-3) (Figs. 1 and 2, Supplemental Table S2). Chondroid matrix-producing tumors were nodular with circumscribed borders and composed of carcinoma cells directly embedded within cartilaginous-like matrix without an associated spindle cell component (Fig. 1 and Supplemental Table S3) [5]. Pure spindle cell carcinomas were entirely sarcomatoid and lacked conventional ductal, squamous, or other heterologous elements. All tumors designated as matrix-producing, squamous, spindled, or squamous/spindled lacked differentiation along other metaplastic lineages. Twenty-one (75%) tumors were associated with a ductal component, including fifteen (54%) with invasive ductal carcinoma and twelve (43%) with DCIS (Table 1 and Supplemental Table S2). Aside from two pure spindle cell carcinomas (Sp2 and Sp3), all tumors showed features of modified Scarff-Bloom-Richardson grade 3. When present, DCIS was high nuclear grade in all but two cases and was predominantly solid and comedo patterns (Table 1). Ninety-three percent (26/28) of tumors were triple negative; one mixed spindle and squamous carcinoma (SS4) was HER2-positive in the squamous but not the spindle cell component, and one squamous cell carcinoma (Sq5) was ER-positive. Nine (32%) tumors had axillary lymph node metastases, of which five (56%) were of mixed squamous and spindle cell or squamous cell histology. Of patients with clinical follow-up (n = 25; mean 31, range 3–167 months), distant metastases were only identified in primary tumors with mixed heterologous components (M1–M3), and all three were metastatic (Table 1).
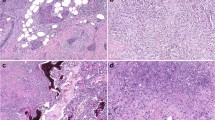
Representative images of metaplastic breast carcinomas. a–c Pure spindle cell carcinomas were entirely composed of spindled tumor cells without other heterologous or conventional ductal elements (Sp1, Sp2, and Sp4, respectively). d–f Metaplastic carcinomas with squamous differentiation (Sq1, Sq3, and Sq4, respectively). g–i Metaplastic carcinomas with mixed squamous and spindle cell differentiation (SS1, SS3, and SS4, respectively). j–l Matrix-producing metaplastic carcinomas with chondroid differentiation (C1, C6, and C7, respectively). Carcinoma cells directly transition to tumor cells embedded within chondroid matrix without associated spindled tumor cells or other heterologous components. m–o Metaplastic carcinomas with mixed heterologous differentiation (M1, M2, and M3, respectively). These tumors demonstrated admixed areas of differentiation along multiple heterologous lineages. Areas containing osseous differentiation were sequenced and are shown here, along with foci of spindled tumor cells (m-o), chondroid (o), and squamous (o) elements. All images hematoxylin and eosin
Genomics of metaplastic carcinomas and correlation with histologic subtype
Lineage differentiation of the sequenced and non-sequenced invasive tumor components is shown in figure 2. The mean target sequencing coverage was 440 (±250) unique reads per target interval (mean 474 ± 256 for invasive cancer, 306 ± 178 for DCIS) (Supplemental Table S4). The number of identified nonsynonymous coding mutations across the 2.8 megabase footprint of the panel was highly variable and ranged from 1 to 51 per invasive tumor (mean 9.2 ± 9.6), without differences across subtypes (Fig. 2 and Supplemental Table S5). The most frequently identified pathogenic aberrations were in PI-3 kinase pathway genes and in TP53, which were each altered in 64% (18/28) of tumors. Sixty-one percent (17/28) of all tumors harbored either PIK3CA activating mutations/amplification or PIK3R1 inframe indels/truncating mutations, and this was dependent on histologic subtype (Figs. 2 and 3). No chondroid matrix-producing tumors harbored mutations in PIK3CA or PIK3R1 or in other PI-3 kinase pathway genes (p < 0.001 vs. other subtypes). In contrast, all other (squamous, squamous/spindled, spindle cell, and mixed) metaplastic carcinomas had PI-3 kinase pathway aberrations, including mutations in PIK3CA or PIK3R1 in 17 of the 18 (94%) tumors. The only non-matrix-producing tumor lacking mutations in PIK3CA or PIK3R1 (squamous cell carcinoma Sq5) harbored an activating mutation (p.E17K) in the downstream kinase AKT1 (Fig. 2 and Supplemental Table S5). Pathogenic Ras-Mitogen activated protein (Map) kinase pathway aberrations were identified in 25% (7/28) of tumors and included activating variants of KRAS (n = 2) and HRAS (n = 2), deep deletions of NF1 (n = 2), and a MAP2K1 missense hotspot mutation (Figs. 2 and 3, Supplemental Table S5). Similar to PI-3 kinase pathway aberrations, Ras-Map kinase pathway mutations were not identified in chondroid matrix-producing tumors (0/10 vs. 7/18 [39%] other subtypes, p = 0.03) (Figs. 2 and 3). Hotspot TERT promoter mutations were identified in 39% (7/18) of non-chondroid tumors, including 4/5 spindle cell carcinomas, 2/5 mixed spindled/squamous cell carcinomas, and 1/5 squamous cell carcinomas. The C228T (chr5: 1295228) variant was identified in 6/7 tumors, whereas one squamous cell carcinoma harbored the C250T (chr5: 1295250) variant (Supplemental fig S5). No TERT promoter mutations were identified in chondroid matrix-producing carcinomas (0/10, 0%; p = 0.03) (Figs. 2 and 3). These tumors were instead enriched for TP53 mutations (90%) but were otherwise more heterogeneous, with inactivating mutations in tumor suppressors such as NOTCH1, chromatin remodeling genes (ARID1A, KMT2A, KMT2D), and DNA repair genes (ERCC2, PALB2) (Fig. 2 and Supplemental Table S5). None of the pure spindle cell carcinomas (0/5) harbored TP53 mutations, in contrast to the high frequency (18/23, 78%) identified across the other histologic groups (p = 0.003) (Figs. 2, 3). The genetic profiles of tumors with mixed heterologous differentiation in which the osseous component was sequenced were overall similar to other non-matrix-producing tumors and included pathogenic PIK3CA/PIK3R1 and TP53 mutations in all cases (Figs. 2, 3).
Genetic heterogeneity of metaplastic carcinoma histologic subtypes with comparison to triple negative carcinomas of no special type. a Frequency of genetic pathway aberrations between histologic subtypes. Percentages reflect total relative number of tumors with designated mutation(s). b Comparison of genetic pathway aberrations between metaplastic carcinomas and triple negative carcinomas from the TCGA dataset [45, 46]. TCGA, The Cancer Genome Atlas; TN, triple negative; MCB, metaplastic breast carcinoma; *p < 0.05
Copy number analysis revealed variable numbers of chromosomal copy number gains and losses between and within histologic subgroups, overall ranging from 0 to 51 alterations per tumor (mean 12.6 ± 10.1). Chondroid matrix-producing carcinomas had higher overall numbers of chromosomal copy number changes (mean 23.1 ± 10.9) than the other subtypes (mean 7.7 ± 9.6, p = 0.002), with mixed squamous/spindle cell and pure spindle cell carcinomas each showing few copy number alterations (mean 1.5 ± 3 and 2.8 ± 3.1, respectively; p < 0.005 each vs. chondroid matrix-producing tumors). Tumors with squamous differentiation also showed high numbers of chromosomal alterations (mean 14 ± 9.5, range 0–20), similar to chondroid matrix-producing carcinomas (p = 0.171 vs. chondroid matrix-producing, p = 0.046 vs. squamous/spindle, and p = 0.06 vs. spindle cell carcinomas) (Supplemental Table S6).
Comparing genomic profiles of metaplastic carcinomas with other triple negative breast carcinomas
We compared the genomic profiles of metaplastic carcinomas in our study population to publicly available sequencing data of triple negative breast cancers (n = 125) analyzed in the TCGA (Fig. 3) [45, 46]. The frequency of TP53 mutations was overall comparable between metaplastic carcinomas and TCGA triple negative tumors (64% vs. 58%, respectively), but TP53 mutations were more common in chondroid matrix-producing carcinomas (90%, p = 0.05). Metaplastic carcinomas as a group more frequently harbored PIK3CA (32% vs. 13%, p = 0.021) or PIK3R1 (29% vs. 2%, p < 0.001) mutations than triple negative TCGA tumors, with 61% of metaplastic carcinomas having pathogenic aberrations in either gene, compared to only 14% of TCGA triple negative tumors (p < 0.001). The differences were even more striking when matrix-producing carcinomas were excluded (50% PIK3CA, 44% PIK3R1, and 94% PIK3CA or PIK3R1 mutations in non-matrix-producing tumors, p < 0.001 each vs. TCGA). No significant differences were identified between metaplastic carcinomas and TCGA triple negative tumors in other PI-3 kinase pathway genes, such as PTEN (14% vs. 7%, p = 0.258) or AKT1 (1 vs. 4%, p = 0.334). Metaplastic carcinomas also had more frequent Ras-Map kinase pathway genetic aberrations than TCGA triple negative tumors (25 vs. 7%, respectively, p = 0.005), a difference which was further amplified in non-matrix-producing tumors (39%, p < 0.001). This increased frequency in Ras-Map kinase pathway mutations was due to more activating HRAS or KRAS mutations in metaplastic carcinomas (14% in all tumors, 22% in non-chondroid matrix tumors) compared to TCGA triple negative tumors, in which such mutations are rare (1%, p = 0.004). The results were similar when excluding the rare ER+ (Sq5) and HER2+ (SS4) metaplastic carcinomas from analysis (data not shown).
Genetic relationship of metaplastic carcinomas with paired invasive and in situ ductal carcinoma
Ductal carcinoma was microdissected and sequenced separately from the invasive heterologous components of nine metaplastic carcinomas, including paired DCIS of eight tumors (C6, C9, Sq2, Sq4, Sq5, Sp1, Sp5, and SS3) and paired invasive ductal carcinoma of two tumors with either chondroid matrix-production (C1) or squamous (Sq4) differentiation (Figs. 4, 5). Of the eight tumors in which paired DCIS was sequenced, two were pure spindle cell carcinomas (Sp1 and Sp5), and DCIS in one of these (Sp5) was apocrine type. DCIS in cases of chondroid matrix-producing (C6 and C9), squamous (Sq2, Sq4, and Sq5) or mixed squamous/spindle cell (SS3) carcinomas was of no special histologic type. In all cases, DCIS and invasive heterologous components were found to be genetically related, as determined by shared single nucleotide variants/indels (7/8 cases) and/or shared copy number changes (6/6 cases) (Fig. 4, Supplemental Tables S5 and S6). Pathogenic activating mutations in PI-3 kinase or Ras pathway genes were present in both DCIS and invasive cancer in all paired cases harboring these alterations (n = 6), indicative of an early event in tumorigenesis. In contrast, inactivating mutations in tumor suppressor genes such as RB1 (Sq2), CDKN2A (Sp5), SMAD4 (Sq4), and KMT2D (Sq4) were associated with stromal invasion and/or tumor progression (Fig. 4). The lack of these mutations in DCIS could not be explained by differences in tumor cellularity, depth of coverage, or mutant allele frequencies between the components (Supplemental Tables S4 and S5). In two cases (Sq2 and SS3), shared TP53 mutations were identified in the paired DCIS and invasive components, whereas two other tumors (C6 and Sq4) harbored different TP53 mutations in DCIS and paired invasive cancer, with the DCIS-associated variants present at subclonal mutant allele frequency (Fig. 4). The findings in the latter tumors support intratumoral heterogeneity of TP53-mutant clones in DCIS with outgrowth of a dominant clone upon progression to invasion. In addition to shared copy number changes between the in situ and invasive components in all informative tumors, private copy number changes were also identified in the DCIS of 4/6 cases and in the invasive components of 5/6 cases (Fig. 4).
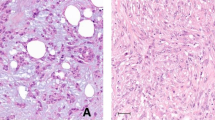
Genomic aberrations in paired metaplastic carcinomas and associated ductal carcinoma in situ. a Summary of pathogenic genomic aberrations in paired invasive and in situ carcinomas. b Representative images of paired ductal carcinoma in situ and invasive metaplastic carcinomas of different histologic subtypes (hematoxylin and eosin). c Representative copy number plots of paired ductal carcinoma in situ and invasive metaplastic carcinoma from tumors with squamous (Sq5) and spindle cell (Sp5) differentiation. Small red arrows highlight shared copy number alterations between in situ and invasive carcinoma, and large black arrows highlight alterations unique to only one of the two components per tumor. DCIS, ductal carcinoma in situ; Inv, invasive; chr, chromosome
Genetic progression of metaplastic carcinomas from ductal to heterologous differentiation. a Paired ductal carcinoma in situ, invasive ductal, and invasive squamous components of tumor Sq4 were microdissected and sequenced separately. Clonal PIK3R1 (p.579_582del), PTEN (p.Y240fs), and ZNF703 (p.179_181del) mutations were shared between all three components. A clonal pathogenic TP53 missense mutation (p.G266R) was shared only between invasive ductal and squamous components, whereas the sampled in situ component harbored two different TP53 missense mutations (p.K132N, p.R213L) at subclonal allele frequencies that were not present in the invasive cancer. Histologic progression to squamous cell carcinoma was associated with unique truncating pathogenic KMT2D (p.S2927*) and SMAD4 (p.Q334*) mutations. b Paired invasive ductal and invasive chondroid components of tumor C1 were microdissected and sequenced separately. FGFR1 and MCL1 amplifications, as well as frameshift NOTCH1 (p.P2514fs), missense TP53 (p.V173M), and missense NT5C2 (p.D306G) mutations, were shared between components. Pathogenic ATRX (p.E873*), RB1 (c.1128-2 A > G), and ARID1AI (p.E1904fs) mutations were unique to the chondroid component, along with numerous additional variants of unknown significance at clonal and subclonal allele frequencies. DCIS, ductal carcinoma in situ
In both tumors in which the invasive ductal and heterologous components were separately sequenced (C1 and Sq4), the paired elements were genetically related to one another, as determined by multiple shared single nucleotide variants/indels and copy number changes (Fig. 5, Supplemental Tables S5 and S6). The chondroid component of tumor C1 and the squamous component of tumor Sq4 each harbored additional clonal pathogenic mutations (in ATRX/RB1/ARID1A and SMAD4/KMT2D, respectively) that were not present in the respective paired ductal carcinoma components, and the number of private variants of unknown significance was higher in heterologous components than paired ductal carcinoma (n = 43 vs. 11 in tumor C1 and n = 11 vs. 4 in tumor Sq4, respectively) (Fig. 5 and Supplemental Table S5). The heterologous components of tumors C1 and Sq4 also harbored more chromosomal copy number changes than paired ductal components (Supplemental Table S6). Analysis of separately sequenced DCIS, invasive ductal, and invasive squamous components in squamous/ductal tumor Sq4 was particularly illustrative for detailing genetic relatedness and tumor progression (Fig. 5). In this tumor, inframe PIK3R1 (p.579_582del) and frameshift PTEN (p.Y240fs) mutations were identified as early events present in all three components, whereas a clonal missense TP53 mutation (p.G266R) was acquired during progression to invasion and shared among the invasive components but not DCIS. The invasive squamous component additionally harbored clonal inactivating SMAD4 (Q334*) and KMT2D (S2927*) mutations, as well as numerous variants of unknown significance not present in either in situ or invasive ductal carcinoma (Fig. 5).
In contrast to the results of metaplastic tumors with ductal components, separately sequenced squamous and spindle cell components of a mixed squamous/spindle cell carcinoma (SS3) showed no pathogenic genetic differences between heterologous components (Fig. 4 and Supplemental Table S5).
Discussion
The clinicopathologic, histopathologic, and molecular heterogeneity of triple negative breast cancers is well established [47,48,49]. We show that metaplastic breast carcinomas comprise a genetically distinct subgroup of triple negative tumors with a high frequency of PI-3 kinase and Ras-Map kinase pathway aberrations compared to other triple negative breast cancers. Previous studies have also demonstrated PI-3 kinase pathway alterations in metaplastic carcinomas but were limited by lack of histologic annotation, inclusion of mixed tumors, inclusion of only sarcomatoid or squamous variants, or analysis of advanced stage treated tumors [18, 22, 23, 26]. Our data indicate that metaplastic carcinomas are genetically heterogeneous and that this correlates broadly with histologic subtype. Chondroid matrix-producing carcinomas conspicuously lack PIK3CA/PIK3R and Ras pathway aberrations, which are very common in other subtypes, including tumors with squamous and/or spindle cell differentiation and tumors with mixed heterologous elements. In this context, our findings expand on those of Ng et al. [24], who also described a lack of PIK3CA mutations in tumors with chondroid metaplasia, although whether the tumors in their study were strictly defined matrix-producing carcinomas is unclear [5]. We suggest that matrix-producing carcinomas are not only histologically but also genetically distinct from other metaplastic carcinomas, including those with foci of chondroid metaplasia in a mixed heterologous background, supporting classification as a distinct subgroup. Enrichment of PI-3 kinase aberrations only in non-matrix producing carcinomas may have treatment implications, as the demonstrated clinical efficacy of PI-3 kinase inhibitors in metaplastic carcinomas and triple negative tumors of mesenchymal molecular subtype correlates with PI-3 kinase pathway mutations [21, 23]. This raises the consideration that matrix-producing tumors may be less likely than other histologic variants to respond to PI-3 kinase pathway inhibition, which will require further study.
Hotspot TERT promoter mutations, which upregulate telomerase reverse transcriptase (TERT) expression and activate telomerase, are among of the most frequently identified mutations in certain tumors but have been considered to be absent or exceedingly rare in carcinomas of the breast [50,51,52,53]. A recent study addressing this issue identified TERT promoter mutations in only 3 (0.9%) of 319 invasive breast cancers, two of which were ER/PR-positive and one of which was triple negative [53]. Other groups have not found TERT promoter mutations in several breast cancer cohorts [51, 52]. We demonstrate for the first time a high frequency of hotspot TERT promoter mutations in metaplastic breast carcinomas and specifically in tumors with spindle cell and/or squamous differentiation (39%) but not matrix-producing carcinomas, which highlights the importance of telomerase activation in the former subgroups. TERT promoter mutations have been associated with poor clinical outcome in several tumor types [54, 55], and TERT overexpression in breast cancer has been correlated with poor response to chemotherapy [56, 57]. Whether TERT promoter mutations are prognostically significant in metaplastic carcinomas requires further study. Given that these mutations are considered to be among the earliest genetic events in tumorigenesis of other cancers [58, 59], it is interesting that only one of two metaplastic carcinomas with TERT promoter mutation in which both components were analyzed also harbored the mutation in paired high grade DCIS (Sp5), whereas intermediate grade DCIS associated with the other tumor (Sp1) lacked the mutation.
The absence of TP53 mutations in spindle cell carcinomas in our study contrasts with the high frequency in other subtypes and in metaplastic carcinomas in general [24, 26]. Few studies have specifically analyzed the genetics of pure spindle cell metaplastic carcinomas, but both sequenced tumors reported by Ross et al also lacked TP53 mutations [26]. Ng et al. reported TP53 mutations in five of ten tumors in which a spindle cell component was sequenced, but these tumors were annotated to comprise other areas with non-spindle cell elements, including squamous, chondroid, osseous, and epithelioid areas, and so likely represent a heterogeneous group of mixed tumors [26]. Although we cannot definitely exclude small sample size bias in our study and in others [24, 26], it appears that TP53 mutations are at least decreased in frequency if not absent from this subgroup, suggesting distinct pathogenesis from other metaplastic carcinomas. Two of the five tumors in our study were of intermediate nuclear grade; whether this is may be related to lack of TP53 mutations is uncertain. Given that the distinction of metaplastic spindle cell carcinoma from phyllodes tumor with stromal overgrowth is a commonly encountered diagnostic dilemma, our results posit the testable hypothesis that TP53 mutation testing or p53 immunostaining may be useful in this differential diagnosis, as malignant phyllodes tumors frequently harbor TP53 mutations [33, 60]. On the other hand, we show that metaplastic carcinomas, like malignant phyllodes tumors, often harbor PI-3 kinase, Ras-Map kinase, and TERT promoter mutations, which are therefore not diagnostically useful in the differential [33, 60].
The repertoire of somatic mutations in DCIS associated with metaplastic carcinoma and its relationship to the invasive tumor has not been investigated prior to this study. We confirm the monoclonality of DCIS and paired metaplastic carcinoma. The presence of DCIS in association with a squamous, spindle cell, or otherwise mesenchymal-like tumor in the breast is often used as evidence to support a diagnosis of metaplastic carcinoma, and our genetic findings confirm the utility of this approach, regardless of the type of differentiation of the invasive or in situ tumor. Indeed, we show that even a histologically disparate DCIS with apocrine differentiation can be a direct precursor for spindle cell carcinoma.
Studies of conventional DCIS have demonstrated largely similar mutational and copy number profiles between the in situ and invasive components, albeit with increased numbers of copy number alterations in invasive cancer, and our results in metaplastic carcinomas are consistent with this [61,62,63,64,65]. As in conventional DCIS [62, 63, 65, 66], we show that PIK3CA alterations are early events in metaplastic carcinoma, with mutations in this and other growth-promoting PI-3 kinase/Ras-Map kinase pathway genes being present in clonal frequencies of associated DCIS and paired invasive tumors. In contrast, although p53 and Rb inactivation may also be early events in conventional [62, 64, 67,68,69,70] and metaplastic carcinoma-associated DCIS, mutations in TP53, RB1 and CDKN2A were specifically associated with stromal invasion in individual metaplastic carcinomas. These findings support a convergent model of tumor invasion, in which multiple pathways that are specific for individual tumors can be responsible for progression to invasion. A similar model has been proposed for conventional in situ-invasive ductal carcinoma [63, 71]. Our findings thus further support the idea that paired tumor analysis is most useful to uncover putative genetic drivers of invasion and help explain why studies comparing non-matched components have not consistently identified such factors [71,72,73]. These results also highlight the presence of intratumoral heterogeneity early during tumor development, with TP53 subclones, as well as chromosomal gains and losses, present in DCIS but not invasive components of some tumors. We speculate that this likely reflects a bottleneck effect of tumor evolution in selection for invasive clones, as has previously been proposed for conventional ductal carcinomas [63, 71, 74]. Lastly, analysis of paired DCIS, invasive ductal carcinoma, and invasive squamous cell carcinoma in the same tumor (Sq4) highlights the stepwise dysregulation of oncogenic drivers such as TP53, SMAD4, and KMT2D during tumor evolution. The findings support a model in which heterologous components are directly evolved and genetically progressed from ductal carcinoma. Our study thereby confirms previous speculation based on a detailed histologic review of metaplastic carcinomas and further supports a similar conclusion of metaplastic carcinogenesis based on recent genetic analysis [75, 76].
References
Reis-Filho J, Lakhani S, Gobbi H, et al.. Metaplastic carcinoma. In: Lakhani S, Ellis I, Schnitt S, Tan P, van de Vijver M, editors. WHO classification of tumours of the breast. 4th edn. Lyon, France: International Agency for Research on Cancer (IARC); 2012. p. 48–52.
Weigelt B, Eberle C, Cowell CF, et al. Metaplastic breast carcinoma: more than a special type. Nat Rev Cancer. 2014;14:147–8.
Wargotz ES, Deos PH, Norris HJ. Metaplastic carcinomas of the breast. II. Spindle cell carcinoma. Hum Pathol. 1989;20:732–40.
Wargotz ES, Norris HJ. Metaplastic carcinomas of the breast. III. Carcinosarcoma Cancer. 1989;64:1490–9.
Wargotz ES, Norris HJ. Metaplastic carcinomas of the breast. I. Matrix-producing carcinoma. Hum Pathol. 1989;20:628–35.
Chhieng C, Cranor M, Lesser ME, et al. Metaplastic carcinoma of the breast with osteocartilaginous heterologous elements. Am J Surg Pathol. 1998;22:188–94.
Cimino-Mathews A, Verma S, Figueroa-Magalhaes MC, et al. A clinicopathologic analysis of 45 patients with metaplastic breast carcinoma. Am J Clin Pathol. 2016;145:365–72.
El Zein D, Hughes M, Kumar S, et al. Metaplastic carcinoma of the breast is more aggressive than triple-negative breast cancer: a study from a single institution and review of literature. Clin Breast Cancer. 2017;17:382–91.
Jung SY, Kim HY, Nam BH, et al. Worse prognosis of metaplastic breast cancer patients than other patients with triple-negative breast cancer. Breast Cancer Res Treat. 2010;120:627–37.
Luini A, Aguilar M, Gatti G, et al. Metaplastic carcinoma of the breast, an unusual disease with worse prognosis: the experience of the European Institute of Oncology and review of the literature. Breast Cancer Res Treat. 2007;101:349–53.
Schwartz TL, Mogal H, Papageorgiou C, et al. Metaplastic breast cancer: histologic characteristics, prognostic factors and systemic treatment strategies. Exp Hematol Oncol. 2013;2:31.
Zhang Y, Lv F, Yang Y, et al. Clinicopathological features and prognosis of metaplastic breast carcinoma: experience of a Major Chinese Cancer Center. PLoS One. 2015;10:e0131409.
Song Y, Liu X, Zhang G, et al. Unique clinicopathological features of metaplastic breast carcinoma compared with invasive ductal carcinoma and poor prognostic indicators. World J Surg Oncol. 2013;11:129.
Rakha EA, Tan PH, Varga Z, et al. Prognostic factors in metaplastic carcinoma of the breast: a multi-institutional study. Br J Cancer. 2015;112:283–9.
Tzanninis IG, Kotteas EA, Ntanasis-Stathopoulos I, et al. Management and outcomes in metaplastic breast cancer. Clin Breast Cancer. 2016;16:437–43.
Weigelt B, Kreike B, Reis-Filho JS. Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res Treat. 2009;117:273–80.
Weigelt B, Ng CK, Shen R, et al. Metaplastic breast carcinomas display genomic and transcriptomic heterogeneity [corrected]. Mod Pathol. 2015;28:340–51.
Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–24.
Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68.
Piscuoglio S, Ng CKY, Geyer FC, et al. Genomic and transcriptomic heterogeneity in metaplastic carcinomas of the breast. NPJ Breast Cancer. 2017;3:48.
Basho RK, Gilcrease M, Murthy RK, et al. Targeting the PI3K/AKT/mTOR pathway for the treatment of mesenchymal triple-negative breast cancer: evidence from a phase 1 trial of mTOR inhibition in combination with liposomal doxorubicin and bevacizumab. JAMA Oncol. 2017;3:509–15.
Joneja U, Vranic S, Swensen J, et al. Comprehensive profiling of metaplastic breast carcinomas reveals frequent overexpression of programmed death-ligand 1. J Clin Pathol. 2017;70:255–9.
Moulder S, Helgason T, Janku F, et al. Inhibition of the phosphoinositide 3-kinase pathway for the treatment of patients with metastatic metaplastic breast cancer. Ann Oncol. 2015;26:1346–52.
Ng CKY, Piscuoglio S, Geyer FC, et al. The landscape of somatic genetic alterations in metaplastic breast carcinomas. Clin Cancer Res. 2017;23:3859–70.
Reis-Filho JS, Pinheiro C, Lambros MB, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445–53.
Ross JS, Badve S, Wang K, et al. Genomic profiling of advanced-stage, metaplastic breast carcinoma by next-generation sequencing reveals frequent, targetable genomic abnormalities and potential new treatment options. Arch Pathol Lab Med. 2015;139:642–9.
Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220:562–73.
Lien HC, Lin CW, Mao TL, et al. p53 overexpression and mutation in metaplastic carcinoma of the breast: genetic evidence for a monoclonal origin of both the carcinomatous and the heterogeneous sarcomatous components. J Pathol. 2004;204:131–9.
Teixeira MR, Qvist H, Bohler PJ, et al. Cytogenetic analysis shows that carcinosarcomas of the breast are of monoclonal origin. Genes Chromosomes Cancer. 1998;22:145–51.
Wang X, Mori I, Tang W, et al. Metaplastic carcinoma of the breast: p53 analysis identified the same point mutation in the three histologic components. Mod Pathol. 2001;14:1183–6.
Zhuang Z, Lininger RA, Man YG, et al. Identical clonality of both components of mammary carcinosarcoma with differential loss of heterozygosity. Mod Pathol. 1997;10:354–62.
Krings G, Joseph NM, Bean GR, et al. Genomic profiling of breast secretory carcinomas reveals distinct genetics from other breast cancers and similarity to mammary analog secretory carcinomas. Mod Pathol. 2017;30:1086–99.
Liu SY, Joseph NM, Ravindranathan A, et al. Genomic profiling of malignant phyllodes tumors reveals aberrations in FGFR1 and PI-3 kinase/RAS signaling pathways and provides insights into intratumoral heterogeneity. Mod Pathol. 2016;29:1012–27.
DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–95.
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Talevich E, Shain AH, Botton T, et al. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12:e1004873.
Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11 0 1–33.
Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66.
Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71.
Rausch T, Zichner T, Schlattl A, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–9.
Garrison E, Marth G. Haplotype-Based Var Detect Short-Read Seq arXiv. 2012;1207:3907. [q-bio.GN]
Picard: A set of tools (in Java) for working with next generation sequencing data in the BAM: Broad Institute. http://broadinstitute.github.io/picard.
Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Mayer IA, Abramson VG, Lehmann BD, et al. New strategies for triple-negative breast cancer--deciphering the heterogeneity. Clin Cancer Res. 2014;20:782–90.
Metzger-Filho O, Tutt A, de Azambuja E, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–87.
Pareja F, Geyer FC, Marchio C, et al. Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer. 2016;2:16036.
Bell RJ, Rube HT, Xavier-Magalhaes A, et al. Understanding TERT promoter mutations: a common path to immortality. Mol Cancer Res. 2016;14:315–23.
Gay-Bellile M, Veronese L, Combes P, et al. TERT promoter status and gene copy number gains: effect on TERT expression and association with prognosis in breast cancer. Oncotarget. 2017;8:77540–51.
Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110:6021–6.
Shimoi T, Yoshida M, Kitamura Y, et al. TERT promoter hotspot mutations in breast cancer. Breast Cancer. 2017;25:292–6.
George JR, Henderson YC, Williams MD, et al. Association of TERT promoter mutation, but not BRAF mutation, with increased mortality in PTC. J Clin Endocrinol Metab. 2015;100:E1550–9.
Griewank KG, Murali R, Puig-Butille JA, et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst 2014;106.dju246
Gay-Bellile M, Romero P, Cayre A, et al. ERCC1 and telomere status in breast tumours treated with neoadjuvant chemotherapy and their association with patient prognosis. J Pathol Clin Res. 2016;2:234–46.
Lu L, Zhang C, Zhu G, et al. Telomerase expression and telomere length in breast cancer and their associations with adjuvant treatment and disease outcome. Breast Cancer Res. 2011;13:R56.
Kinde I, Munari E, Faraj SF, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73:7162–7.
Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218.
Piscuoglio S, Ng CK, Murray M, et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression. J Pathol. 2016;238:508–18.
Pang JM, Gorringe KL, Wong SQ, et al. Appraisal of the technologies and review of the genomic landscape of ductal carcinoma in situ of the breast. Breast Cancer Res. 2015;17:80.
Pang JB, Savas P, Fellowes AP, et al. Breast ductal carcinoma in situ carry mutational driver events representative of invasive breast cancer. Mod Pathol. 2017;30:952–63.
Hernandez L, Wilkerson PM, Lambros MB, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J Pathol. 2012;227:42–52.
Kim SY, Jung SH, Kim MS, et al. Genomic differences between pure ductal carcinoma in situ and synchronous ductal carcinoma in situ with invasive breast cancer. Oncotarget. 2015;6:7597–607.
Abba MC, Gong T, Lu Y, et al. A molecular portrait of high-grade ductal carcinoma in situ. Cancer Res. 2015;75:3980–90.
Miron A, Varadi M, Carrasco D, et al. PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res. 2010;70:5674–8.
Kang JH, Kim SJ, Noh DY, et al. The timing and characterization of p53 mutations in progression from atypical ductal hyperplasia to invasive lesions in the breast cancer. J Mol Med. 2001;79:648–55.
Zhou W, Muggerud AA, Vu P, et al. Full sequencing of TP53 identifies identical mutations within in situ and invasive components in breast cancer suggesting clonal evolution. Mol Oncol. 2009;3:214–9.
Vincent-Salomon A, Lucchesi C, Gruel N, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:1956–65.
Subhawong AP, Subhawong T, Nassar H, et al. Most basal-like breast carcinomas demonstrate the same Rb-/p16 + immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol. 2009;33:163–75.
Cowell CF, Weigelt B, Sakr RA, et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol Oncol. 2013;7:859–69.
Jang M, Kim E, Choi Y, et al. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14:R115.
Robanus-Maandag EC, Bosch CA, Kristel PM, et al. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. J Pathol. 2003;201:75–82.
Martelotto LG, Baslan T, Kendall J, et al. Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat Med. 2017;23:376–85.
van Deurzen CH, Lee AH, Gill MS, et al. Metaplastic breast carcinoma: tumour histogenesis or dedifferentiation? J Pathol. 2011;224:434–7.
Avigdor BE, Beierl K, Gocke CD, et al. Whole-exome sequencing of metaplastic breast carcinoma indicates monoclonality with associated ductal carcinoma component. Clin Cancer Res. 2017;23:4875–84.
Acknowledgements
We thank the UCSF Clinical Cancer Genomic Laboratory.
Funding
This study was funded by the Department of Pathology of the University of California San Francisco.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Krings, G., Chen, YY. Genomic profiling of metaplastic breast carcinomas reveals genetic heterogeneity and relationship to ductal carcinoma. Mod Pathol 31, 1661–1674 (2018). https://doi.org/10.1038/s41379-018-0081-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0081-z
This article is cited by
-
Regulation and clinical potential of telomerase reverse transcriptase (TERT/hTERT) in breast cancer
Cell Communication and Signaling (2023)
-
Transcriptomic alterations underlying metaplasia into specific metaplastic components in metaplastic breast carcinoma
Breast Cancer Research (2023)
-
The mixed subtype has a worse prognosis than other histological subtypes: a retrospective analysis of 217 patients with metaplastic breast cancer
Breast Cancer Research and Treatment (2023)
-
Genomic characterization and tumor evolution in paired samples of metaplastic breast carcinoma
Modern Pathology (2022)
-
TERT promoter hotspot mutations and gene amplification in metaplastic breast cancer
npj Breast Cancer (2021)