Abstract
Basal cell salivary neoplasms display similar cyto-morphologic features and are classified into adenoma and adenocarcinoma based on the presence or absence of tumor invasion at diagnosis. These neoplasms also share considerable phenotypic resemblance and co-exist with certain dermal adnexal tumors harboring the CYLD gene mutations inferring common genetic association. We sequenced the CYLD gene in both basal cell adenomas and adenocarcinomas and correlated the findings with CYLD, NF-κB, and β-catenin expression levels and clinicopathologic factors. Twenty mutations were identified and comprised of 3 synonymous and 17 non-synonymous (missense) types involving the coding exons of the CYLD gene. Mutations in exons 9–11 were identified in both adenomas and adenocarcinomas, while mutations in exons 12–20, encoding the USP domain, were exclusively found in carcinomas. Although no significant correlation between CYLD mutations and expression levels of CYLD, NF-κB, and β-catenin or clinicopathologic parameters was found, basal cell adenocarcinomas with multiple mutations showed reduction in CYLD protein expression and pursued aggressive clinical behavior. Our study revealed high incidence and sequential CYLD mutations in both basal cell adenoma and adenocarcinoma supporting a single neoplastic continuum for their evolution and provides evidence for potential diagnostic and therapeutic utility.
Similar content being viewed by others
Introduction
Basal cell salivary neoplasms encompass benign and malignant forms that display uniform basal cell composition in variable morphologic organizations. Given these overlapping features, they are classified into adenoma and adenocarcinoma based on the state of invasion at diagnosis [1,2,3,4]. These characteristics could lead to presurgical and/or post-surgical diagnostic and management challenges. We hypothesize that the morphologic equivalence and co-occurrence of adenoma and adenocarcinoma in some patients sustain the concept of a single neoplastic continuum for their development and progression [5,6,7,8]. Understanding the development of these tumors, however, is hampered by the paucity of molecular genetic information and the lack of surrogate models [9, 10]. Interestingly, basal cell salivary tumors share considerable morphologic and biological characteristics with certain dermal adnexal tumors with CYLD gene mutations [11,12,13,14,15,16,17,18]. In an earlier study, we reported frequent loss of heterozygosity (LOH) at chromosome 16q region and postulated the involvement of CYLD gene in concurrent salivary and dermal basal cell neoplasms [19]. This contention has gained further credence with the finding of CYLD alteration in a patient with dermal-like basal cell salivary tumors [20].
The CYLD gene, mapped to chromosome 16q 12–13 region, spans 56 kb and encodes for protein with deubiquiting enzyme function [14, 21, 22]. The CYLD protein consists of a cytoskeletal-associated proline-glycine, a conserved proline rich, and a catalytic of ubiquitin carboxyl-terminal hydrolase domains. The carboxyl-terminal (USP) domain specifically removes the K-63-linked polyubiquitin chain resulting in the deubiquitination of key cell survival regulatory proteins. Consequently, alteration of the CYLD can lead to sustained activation and deregulation of cell survival proteins associated with tumorigenesis [21, 23,24,25,26,27,28]. Although mutation is the most common CYLD alteration, evidence for its downregulation and activation by genomic and non-genomic modifications have been reported [27,28,29,30,31,32]. Interestingly, no mutation has been reported to involve exons 1–5 of the N-terminal domain of the CYLD gene, suggesting a critical role for this region in the stability of the gene in both wild and mutated states [21, 22, 33]. Currently, the majority of CYLD mutations reported in epithelial malignancies have been confined to exons 9–20 and limited to frameshift and nonsense types, implicating disruption of the deubiquitination domain in tumorigenesis [28, 30, 33].
We posit that CYLD gene alterations underlie the development and progression of salivary basal cell tumors and that these alterations may have an important diagnostic and clinical implications. To test this hypothesis, CYLD gene was sequenced in a series of salivary basal cell adenomas and adenocarcinomas and correlated the findings with the expression of selected downstream effector and the clinicopathologic pathology factors.
Materials and methods
Forty five basal cell salivary neoplasms, 21 (12 adenomas and 9 adenocarcinomas) from Instituto Português de Oncologia de Lisboa Francisco Gentil, Lisboa and 24 (2 adenomas and 22 adenocarcinomas) from MD Anderson Cancer Center, were assessed in this study. Four tumors from the two siblings were also tested [19]. Eight normal parotid gland specimens comprised the materials for the study. The study was approved by the Institutional Board of the respective institutions. Hematoxylin and eosin-stained sections of tumors from both institutions were independently re-reviewed at MD Anderson by a Head and Neck pathologist. Clinicopathologic characteristics were extracted from the patients’ charts, pathologic reports, and review of the available slides.
DNA extraction
DNA extracted from both formalin-fixed paraffin-embedded and available fresh frozen tissues were processed using the Gentra Puregene Tissue Kit (QIAGEN) according to the manufacturer’s protocol. DNA was quantified using thee Nanodrop spectrophotometer (Nanodrop). DNA integrity was analyzed using agarose gel electrophoresis.
CYLD gene sequencing
Mutation analysis for exons 9–20 of the CYLD gene was performed using Sanger sequencing. For PCR primers, see Supplementary table S1. PCR fragments were purified by ExoSAP-IT (Affimetrix) and analyzed using Applied Biosystems 3730X1 DNA analyzer at GENEWIZ, Inc. (New Jersey, USA) and Sequencing and Microarray Facility at MD Anderson Cancer Center.
Mutation classification
All sequence data were manually checked by ABI viewing program. The following definitions were applied to mutation classification: (1) non-synonymous mutation: missense mutations that alter amino acid sequence, (2) synonymous mutations: mutations with no amino acid substitution. Common variants (single-nucleotide polymorphisms) were filtered (dbSNP databases, version 148) in mutational calling.
Immunohistochemistry (IHC)
IHC was performed on 4 µm thin sections of selected paraffin blocks using Autostainer Link 48 (Dako) according to the manufacturer’s instructions. Anti-CYLD (Abcam, ab137524), anti-nuclear factor-κB (anti-NF-κB; Cell signaling, D14E12), and anti-β-Catenin (Dako, Clone: β-Catenin-1) antibodies were used with appropriate antigen retrieval and dilutions. CYLD, NF-κB, and β-Catenin protein expression levels were evaluated for nuclear and/or cytoplasmic staining based on the following criteria: 3+, strong staining in >10% tumor cells; 2+, moderate staining in >10%; 1+, weak staining in >10%; 0, negative or <10% staining. For categorical classification, expression was scored as high (strong and moderate) and low (weak and negative) staining.
Results
Histopathology
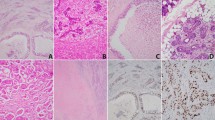
Both adenomas and adenocarcinomas were composed of uniform basal cells forming variable morphologic structures with and without dense eosinophilic basement membrane materials (Fig. 1). Both the inner ductal and outer base cells displayed uniform round-to-oval nuclei with nuclear-to-cytoplasmic ratio. Occasionally, slight cellular pleomorphism and rare mitosis were formed in few adenocarcinomas.
Cellular resemblance of both basal cell adenoma and adenocarcinoma of salivary gland. (A, B) A photomicrograph of basal cell salivary adenoma, note the well-defined tumor and host tissue boundaries. (C, D) A contrasting micrograph of basal cell salivary adenocarcinoma with invasive tumor nests into surrounding soft tissue
Table 1 presents the clinicopathologic parameters of patients with basal cell tumors. Histopathologically, tumors were classified into 11 adenomas and 34 adenocarcinomas. Patients included 22 males and 23 females who ranged in age from 18 to 85 years with a median of 53 years. All tumors were located in the parotid gland. Tumor sizes ranged from 1.5 to 6.0 cm (mean 2.0 cm).
CYLD mutation
Figure 2 illustrates CYLD mutated locations in basal cell salivary neoplasms, and supplementary tables S2 and S3 present the results of mutation analysis in this study. A total of 23 CYLD nucleotide substitutions were identified in 23 of the 45 tumors (51%). Three alterations were single-nucleotide polymorphisms (dbSNP database) identical to those found in corresponding in eight matching normal tissue. The remaining 20 mutations comprised of 17 nonsynonymous (missense) and 3 synonymous mutations (Supplementary tables S2 and S3). Both single-nucleotide polymorphisms and synonymous mutations were excluded from the analysis. Seventeen missense mutations were identified in 4 of the 11 adenomas (36%, Table 1) and 10 of the 34 adenocarcinomas (29%), where 3 adenocarcinomas had 2 concurrent CYLD mutations (Supplementary table S2). Only one mutation was identified in each of the four adenomas. The majority of CYLD mutations in adenocarcinomas occurred within the ubiquitin protease domain (exons 12–20, USP domain), while mutations in adenomas were located in exons 9–11 proximal to the USP domain (Fig. 2 and Table 2, p = 0.087). The most common sites of mutations within the USP domain were found in exon 18.
CYLD mutation in salivary basal cell tumors. CYLD protein domain and mutated locations in basal cell adenoma (BCAd, yellow flag) and basal cell adenocarcinoma (BCAC, red flag). Non-synonymous mutations (missense only in this study) were analyzed. Two examples of missense mutations (p.S457N and p.C799S) are represented. Most of the mutations in basal cell adenocarcinoma occurred in USP domain. CAP cytoskeleton-associated protein glycine-rich (CAP-GLY) domain. Phos_site phosphorylation region of CYLD, USP ubiquitin (Ub)-specific protease domain. Adjacent table indicates the result that CYLD mutation occurred frequently in USP domain of basal cell adenocarcinoma (p = 0.087, Table 2)
CYLD mutation in familial cases
Supplementary table S4 presents the CYLD gene mutation in previously published tumors from the two sisters [19]. Interestingly, exact same mutation from the two sisters were found in basal cell adenocarcinoma and were located in the USP domain [c.2839T>C (p.Y947H)]. Also, both trichoepithelioma and basal cell adenocarcinoma in sister #1 retained the exact single-nucleotide polymorphism in exon 18 (p.D804D, rs2066852).
CYLD alterations and expression of selected targets
Tables 3 and 4 present the correlation between CYLD gene alteration and expression of target genes. Both nuclear and/or cytoplasmic CYLD protein expression (Fig. 3) was noted in 42 tumors with and without CYLD mutations, and both basal cell adenomas and adenocarcinomas showed variably high CYLD protein expression. Two basal cell adenocarcinomas showed decreased expression of CYLD with this gene mutation. Although not significant, trend association between CYLD mutations and its protein expression was only observed in basal cell adenocarcinomas (p = 0.085, Table 3). A significant association between CYLD mutation status (multiple and/or USP domain) and its low CYLD expression (p = 0.048, Table 4) was noted. Both basal cell adenomas and adenocarcinomas expressed both NF-κB and β-Catenin mainly in the cytoplasm and occasionally in the nuclei (Fig. 3). No significant association between CYLD mutation and the expression levels of NF-κB and β-catenin in both basal cell adenomas and adenocarcinomas was found (Tables 3 and 4).
Expression of CYLD, NF-κB, and β-catenin in salivary basal cell tumors. CYLD expression in an adenoma (POBA02) and adenocarcinoma (MDBC04 and MDBC14) with/without CYLD mutation. MDBC04 (CYLD mutation in USP) showed weak CYLD expression. Note that strong cytoplasmic expression with occasional nuclear staining of CYLD was found. NF-κB and β-catenin expression level status was not affected by CYLD expression status (see also Table 3). Nuclear positive of β-catenin expression was found in both adenoma and adenocarcinoma. BCAd basal cell adenoma, BCAC basal cell adenocarcinoma
Clinicopathologic correlation
Tables 1 and 5 present the clinicopathologic characteristics of the entire and adnocarcinoma cohort, respectively. Compared with adenomas and adenocarcinomas, tumor size showed a trend association with malignant diagnosis (p = 0.079, Table 1). Only significant correlation between perineural invasion and metastasis and malignant diagnosis was found (p = 0.044 and 0.042, respectively, Table 1). No significant correlation between the expression levels of either NF-κB or β-Catenin and any of the clinicopathologic parameter was found (Tables 1 and 5). In patients with basal cell adenocarcinomas, a trend correlation between CYLD mutations and perineural invasion was found (p = 0.061, Table 5). Although only three patients with adenocarcinoma had multiple CYLD mutations, those showed poor outcome (Table 5).
Discussion
Basal cell salivary neoplasms encompass benign and malignant forms and share common cellular and architectural manifestations. These pathmorphologic resemblances along with their coincident occurrence in some patients infer sequential neoplastic evolution for their development [1,2,3,4,5,6,7,8]. We postulate that basal cell salivary neoplasms evolve as a single neoplastic continuum with an initial adenoma stage and a subsequent malignant state with temporal progression. Identifying driver genetic events underlying basal cell salivary tumor development therefore is fundamental to early diagnosis and predicting progressive disease. We report, for the first time, frequent and differential domain mutations of the CYLD gene in benign and malignant basal cell salivary gland tumors. The distribution of the mutations were confined to two distinct clusters: one at exons 9–11 that were found in both adenomas and some carcinomas, and the other at exons 12–20 that were exclusively found in carcinomas, suggesting a sequential acquisition with tumor progression [27, 30,31,32].
A notable hallmark in our study is the restricted CYLD gene mutations to the missense type in both benign and malignant forms of basal cell salivary neoplasms. This finding is at variance in its frequency and extent from those reported in dermal adnexal tumors, suggesting tumor context specificity [33]. Interestingly, however, an association between CYLD missense mutations and specific dermal adnexal phenotype has been described [33,34,35]. Together, these findings suggest that CYLD missense mutation may play a role in the genotype–phenotype determination of same forms of these tumors. Our results also advance a potential diagnostic and biological implications for the CYLD mutations where mutational screening by non-invasive presurgical sampling of primary or recurrent tumors could be a better management of certain patients [33]. In this cohort, although no significant correlation between CYLD mutations and patients’ outcome was found, we observed that carcinomas with multiple mutations pursued more aggressive clinical course. Large cohort with long follow-up duration of patients with basal cell salivary neoplasms is needed to validate our findings.
Our results indicate the retention of one allele of the CYLD gene in basal cell salivary neoplasms as evidenced by the persistent CYLD expression in tumors with mutations and argue against the requirement for inactivation of both alleles of CYLD as a tumor-suppressor gene in these tumors [12,13,14, 28]. Similar findings have been reported in familial dermal tumors where no LOH was found in tumors with mutation of the CYLD gene [12, 34,35,36]. Together, these findings suggest that haploinsufficiency and/or dominant-negative isoform effect [37,38,39,40] result in dysregulation of downstream effectors of cellular homeostasis and contribute to basal cell salivary and dermal adnexal tumorigenesis. Accordingly, total loss of the CYLD gene would be incompatible with basal cell tumor development and that either LOH or mutations of the CYLD underlie the development of majority of basal cell salivary tumors.
In our study, CYLD mutations were found in 50% of tumors, suggesting that epigenetic modifications, LOH at chromosome 16q13 region, and/or other alterations including different chromosomal sites could also be associated with basal cell salivary neoplasm development. Notably, alterations of chromosomes 1q22 and 9p21 regions have been reported in dermal adnexal tumors lacking chromosome 16q12-q13 abnormalities, supporting intertumoral and genetic variability [41, 42]. Similarly, alterations in chromosomes 1q and 9p regions have also been identified in multiple basal cell salivary tumors underscoring the kinship of these tumors [43,44,45]. These findings along with evidence for the involvement of PTCH1 gene alterations in these tumors suggest that alternative events, exclusive of CYLD gene alterations, could play a role in basal cell salivary tumorigenesis [46]. Other non-genetic modifications leading to downregulation of the CYLD expression or through the Wnt and NOTCH signaling pathways could also contribute to tumorigenesis [47,48,49,50,51,52].
Importantly, our findings highlight a potential diagnostic and therapeutic implications for CYLD mutational analysis in patients with these basal cell salivary tumors. Diagnostically, mutational screening of CYLD, similar to dermal syndromes, could lead to early classification and stratification of patient for targeted therapy. Recent evidence for an association between CYLD somatic mutations and impairment of the tropomyosin kinase signaling provides an attractive therapeutic target for patients with dermal and basal cell salivary tumors [53, 54]. This is further sustained by new data demonstrating singnifcant inhibitory effect of Lestaurtinib on colony formation and tumor cell proliferation in vitro [53]. The availability of these small molecule agents may be employed in future trials of patients with advanced basal cell salivary adenocarcinomas. Further investigations of surrogate models of these tumors may provide a proof of principle for the application of these agents in patients with recurrent and/or metastatic basal cell salivary adenocarcinoma.
In conclusion, our findings support our hypothesis that basal cell salivary neoplasms represent a sequential neoplastic continuum and that mutation of the CYLD may allow for early classification and the development of future targeted therapeutic modalities. We also maintain that alterations affecting cell death and senescence pathways underlie the indolent nature of salivary basal cell tumorigenesis.
References
El-Naggar AK, Chan JKC, Takata T, et al. Thefourth edition of the head and neck World Health Organization blue book: editors’ perspectives. Hum Pathol. 2017;66:10–12.
Batsakis JG, Luna MA. Basaloid salivary carcinoma. Ann Otol Rhinol Laryngol. 1991;100:785–7.
Batsakis JG, Luna MA, El-Naggar AK. Basaloid monomorphic adenomas. Ann Otol Rhinol Laryngol. 1991;100:687–90.
Wilson TC, Robinson RA. Basal cell adenocarcinoma and Basal cell adenoma of the salivary glands: a clinicopathological review of seventy tumors with comparison of morphologic features and growth control indices. Head Neck Pathol. 2015;9:205–13.
Chen KT. Carcinoma arising in monomorphic adenoma of the salivary gland. Am J Otolaryngol. 1985;6:39–41.
Luna MA, Batsakis JG, Tortoledo ME, et al. Carcinomas ex monomorphic adenoma of salivary glands. J Laryngol Otol. 1989;103:756–9.
Muller S, Barnes L. Basal cell adenocarcinoma of the salivary glands. Report of seven cases and review of the literature. Cancer. 1996;78:2471–7.
Nagao T, Sugano I, Ishida Y, et al. Basal cell adenocarcinoma of the salivary glands - comparison with basal cell adenoma through assessment of cell proliferation, apoptosis, and expression of p53 and bcl-2. Cancer. 1998;82:439–47.
Jo VY, Sholl LM, Krane JF. Distinctive patterns of CTNNB1 (beta-catenin) alterations in salivary gland basal cell adenoma and basal cell adenocarcinoma. Am J Surg Pathol. 2016;40:1143–50.
Wilson TC, Ma D, Tilak A, et al. Next-generation sequencing in salivary gland basal cell adenocarcinoma and basal cell adenoma. Head Neck Pathol. 2016;10:494–500.
Antonescu CR, Terzakis JA. Multiple malignant cylindromas of skin in association with basal cell adenocarcinoma with adenoid cystic features of minor salivary gland. J Cutan Pathol. 1997;24:449–53.
Biggs PJ, Chapman P, Lakhani SR, et al. The cylindromatosis gene (cyld1) on chromosome 16q may be the only tumour suppressor gene involved in the development of cylindromas. Oncogene. 1996;12:1375–7.
Biggs PJ, Wooster R, Ford D, et al. Familial cylindromatosis (turban tumour syndrome) gene localised to chromosome 16q12-q13: evidence for its role as a tumour suppressor gene. Nat Genet. 1995;11:441–3.
Bignell GR, Warren W, Seal S, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–5.
Saggar S, Chernoff KA, Lodha S, et al. CYLD mutations in familial skin appendage tumours. J Med Genet. 2008;45:298–302.
Young AL, Kellermayer R, Szigeti R, et al. CYLD mutations underlie Brooke-Spiegler, familial cylindromatosis, and multiple familial trichoepithelioma syndromes. Clin Genet. 2006;70:246–9.
Zhang XJ, Liang YH, He PP, et al. Identification of the cylindromatosis tumor-suppressor gene responsible for multiple familial trichoepithelioma. J Invest Dermatol. 2004;122:658–64.
Kakagia D, Alexiadis G, Kiziridou A, et al. Brooke-Spiegler syndrome with parotid gland involvement. Eur J Dermatol. 2004;14:139–41.
Choi HR, Batsakis JG, Callender DL, et al. Molecular analysis of chromosome 16q regions in dermal analogue tumors of salivary glands: a genetic link to dermal cylindroma? Am J Surg Pathol. 2002;26:778–83.
Kazakov DV, Benkova K, Michal M, et al. Skin type spiradenoma of the parotid gland with malignant transformation: report of a case with analysis of the CYLD gene. Hum Pathol. 2009;40:1499–1503.
Komander D, Lord CJ, Scheel H, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–64.
Saito K, Kigawa T, Koshiba S, et al. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure. 2004;12:1719–28.
Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–30.
Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–44.
Massoumi R, Chmielarska K, Hennecke K, et al. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–77.
Urbanik T, Kohler BC, Boger RJ, et al. Down-regulation of CYLD as a trigger for NF-kappaB activation and a mechanism of apoptotic resistance in hepatocellular carcinoma cells. Int J Oncol. 2011;38:121–31.
Massoumi R. Ubiquitin chain cleavage: CYLD at work. Trends Biochem Sci. 2010;35:392–9.
Massoumi R, Paus R. Cylindromatosis and the CYLD gene: new lessons on the molecular principles of epithelial growth control. Bioessays. 2007;29:1203–14.
Yang Y, Zhou J. CYLD - a deubiquitylase that acts to fine-tune microtubule properties and functions. J Cell Sci. 2016;129:2289–95.
Blake PW, Toro JR. Update of cylindromatosis gene (CYLD) mutations in Brooke-Spiegler syndrome: novel insights into the role of deubiquitination in cell signaling. Hum Mutat. 2009;30:1025–36.
Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34.
Trompouki E, Hatzivassiliou E, Tsichritzis T, et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–6.
Nagy N, Farkas K, Kemeny L, et al. Phenotype-genotype correlations for clinical variants caused by CYLD mutations. Eur J Med Genet. 2015;58:271–8.
Bowen S, Gill M, Lee DA, et al. Mutations in the CYLD gene in Brooke-Spiegler syndrome, familial cylindromatosis, and multiple familial trichoepithelioma: lack of genotype-phenotype correlation. J Invest Dermatol. 2005;124:919–20.
Poblete Gutierrez P, Eggermann T, Holler D, et al. Phenotype diversity in familial cylindromatosis: a frameshift mutation in the tumor suppressor gene CYLD underlies different tumors of skin appendages. J Invest Dermatol. 2002;119:527–31.
Thomson SA, Rasmussen SA, Zhang J, et al. A new hereditary cylindromatosis family associated with CYLD1 on chromosome 16. Hum Genet. 1999;105:171–3.
Inoue K, Fry EA. Haploinsufficient tumor suppressor genes. Adv Med Biol. 2017;118:83–122.
Kwabi-Addo B, Giri D, Schmidt K, et al. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc Natl Acad Sci USA. 2001;98:11563–8.
Philipp-Staheli J, Payne SR, Kemp CJ. p27(Kip1): regulation and function of a haploinsufficient tumor suppressor and its misregulation in cancer. Exp Cell Res. 2001;264:148–68.
Zurawel RH, Allen C, Wechsler-Reya R, et al. Evidence that haploinsufficiency of Ptch leads to medulloblastoma in mice. Genes Chromosomes Cancer. 2000;28:77–81.
Bare JW, Lebo RV, Epstein EH Jr.. Loss of heterozygosity at chromosome 1q22 in basal cell carcinomas and exclusion of the basal cell nevus syndrome gene from this site. Cancer Res. 1992;52:1494–8.
Harada H, Hashimoto K, Ko MS. The gene for multiple familial trichoepithelioma maps to chromosome 9p21. J Invest Dermatol. 1996;107:41–43.
Saluja K, Rao PH, Myers JN, et al. Novel t(1;3)(q21, p21) translocation in a basal cell adenocarcinoma of the parotid gland: potential association with tumorigenesis. Hum Pathol. 2016;54:189–92.
Hrynchak M, White V, Berean K, et al. Cytogenetic findings in seven lacrimal gland neoplasms. Cancer Genet Cytogenet. 1994;75:133–8.
Toida M, Balazs M, Mori T, et al. Analysis of genetic alterations in salivary gland tumors by comparative genomic hybridization. Cancer Genet Cytogenet. 2001;127:34–37.
Pan S, Dong Q, Sun LS, et al. Mechanisms of inactivation of PTCH1 gene in nevoid basal cell carcinoma syndrome: modification of the two-hit hypothesis. Clin Cancer Res. 2010;16:442–50.
Stegmeier F, Sowa ME, Nalepa G, et al. The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci USA. 2007;104:8869–74.
Sun L, Gao J, Huo L, et al. Tumour suppressor CYLD is a negative regulator of the mitotic kinase Aurora-B. J Pathol. 2010;221:425–32.
Wickstrom SA, Masoumi KC, Khochbin S, et al. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–44.
Tauriello DV, Haegebarth A, Kuper I, et al. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37:607–19.
Hayashi M, Jono H, Shinriki S, et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res Treat. 2014;143:447–57.
Espinosa L, Cathelin S, D’Altri T, et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell. 2010;18:268–81.
Rajan N, Elliott R, Clewes O, et al. Dysregulated TRK signalling is a therapeutic target in CYLD defective tumours. Oncogene. 2011;30:4243–60.
Cranston A, Stocken DD, Stamp E, et al. Tropomyosin Receptor Antagonism in Cylindromatosis (TRAC), an early phase trial of a topical tropomyosin kinase inhibitor as a treatment for inherited CYLD defective skin tumours: study protocol for a randomised controlled trial. Trials. 2017;18:111.
Acknowledgements
The authors are grateful to Ms. Deborah A. Rodriguez and Ms. Cynthia F. Steward for material retrieval and follow-up information and Ms. Jie Li, Ms. Cynthia R Mandel, and Ms. Sheila Fontenot for transcribing and administrative assistance.
Funding
The study is supported in part by the NIH National Institute of Dental and Craniofacial Research and the NIH Office of Rare Diseases Research grant number U01DE019765, the SGTB (Salivary Gland Tumor Biorepository, HHSN268200900039C 04), the Head and Neck SPORE Program grant number P50CA097007, R21DE023656, The Kenneth D. Muller professorship, and NCI Grant P30CA016672.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
M Rito and Y Mitani contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Rito, M., Mitani, Y., Bell, D. et al. Frequent and differential mutations of the CYLD gene in basal cell salivary neoplasms: linkage to tumor development and progression. Mod Pathol 31, 1064–1072 (2018). https://doi.org/10.1038/s41379-018-0018-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0018-6
This article is cited by
-
CYLD induces high oxidative stress and DNA damage through class I HDACs to promote radiosensitivity in nasopharyngeal carcinoma
Cell Death & Disease (2024)
-
Proceedings of the 2024 North American Society of Head and Neck Pathology Companion Meeting, Baltimore, MD, March 24, 2024: Navigating Ancillary Studies in Basaloid/Blue Salivary Tumors
Head and Neck Pathology (2024)
-
Basal Cell Adenoma and Basal Cell Adenocarcinoma: a 50-year Experience From a Single Institution
Head and Neck Pathology (2022)
-
Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Familial Tumor Syndromes
Head and Neck Pathology (2022)
-
The evolving role of molecular pathology in the diagnosis of salivary gland tumours with potential pitfalls
European Archives of Oto-Rhino-Laryngology (2022)