Abstract
Exosomes are cell-derived nanovesicles that have recently gained popularity as potential biomarkers in liquid biopsies due to the large amounts of molecular cargo they carry, such as nucleic acids and proteins. However, most existing exosome-based analytical sensing methods struggle to achieve high sensitivity and high selectivity simultaneously. In this work, we present an electrochemical micro-aptasensor for the highly sensitive detection of exosomes by integrating a micropatterned electrochemical aptasensor and a hybridization chain reaction (HCR) signal amplification method. Specifically, exosomes are enriched on CD63 aptamer-functionalized electrodes and then recognized by HCR products with avidin-horseradish peroxidase (HRP) attached using EpCAM aptamers as bridges. Subsequently, the current signal that is generated through the enzyme reaction between the HRP enzyme and 3,3’,5,5’-tetramethylbenzidine (TMB)/H2O2 directly correlates to the amount of bound HRP on the HCR products and thus to the number of target exosomes. By introducing anti-EpCAM aptamers, micro-aptasensors can detect cancerous exosomes with high specificity. Due to the micropatterned electrodes and HCR dual-amplification strategy, the micro-aptasensors achieve a linear detection response for a wide range of exosome concentrations from 2.5×103 to 1×107 exosomes/mL, with a detection limit of 5×102 exosomes/mL. Moreover, our method successfully detects lung cancer exosomes in serum samples of early-stage and late-stage lung cancer patients, showcasing the great potential for early cancer diagnosis.
Similar content being viewed by others
Introduction
Exosomes, nanosized vesicles with diameters of 30–150 nm, are released by cells during the fusion of multivesicular endosomes with plasma membranes1,2. Exosomes transfer RNA and proteins to recipient cells, thereby facilitating cell-to-cell communication. Recent studies3,4,5 have demonstrated that the expression profiles of exosomal nucleic acids and proteins are altered in many diseases, including cancer, cardiovascular illnesses, infectious diseases, diabetes, neurodegenerative diseases, and depression, demonstrating their promise as a noninvasive biomarker for early detection and diagnosis. For example, many studies show that exosomes secreted by cancer cells carry abundant information about the cells in the form of various biological molecules, such as proteins, lipids, RNA, and DNA fragments6,7,8,9. Some recent studies present evidence that exosomes secreted by cancer cells could contribute to the pathological process and even promote tumor growth, migration, invasion, and chemoresistance10,11,12,13,14. In this regard, exosomes could be an excellent biomarker for early cancer diagnosis, and the concentration of cancer-specific exosomes could be used to determine the corresponding stage of cancer15,16,17.
Great efforts have been devoted to developing methods for exosome analysis and detection based on fluorescence18,19, colorimetry20,21, electrochemistry22,23,24,25,26,27, surface-enhanced Raman scattering (SERS)28,29, surface plasmon resonance (SPR)30,31, photoelectrochemical (PEC)32, and mass spectrometry33. In particular, various electrochemical biosensors have been developed for exosome detection using antibodies as recognition molecules34,35,36,37. One attractive alternative to antibodies is aptamers (i.e., short ssDNA and RNA molecules). Aptamers bind to desired targets with excellent specificity and exhibit merits such as high stability, easy synthesis, and low cost10,38,39,40. Several aptamer-based biosensors have been developed for the determination of exosomes. Li et al. developed an aptasensor based on a hemin/G-quadruplex-assisted signal amplification strategy to perform electrochemical detection of gastric cancer exosomes24. Tan et al. developed an aptamer-based nanotetrahedron-assisted biosensor to capture and detect hepatocellular exosomes41. However, most existing aptamer-based methods are nonspecific target capture methods, which are based on nonspecific markers42 or chemical reactions27. These nonspecific capture methods make it difficult to distinguish between tumor and nontumor exosomes. Therefore, it is highly desirable to develop new aptasensors to overcome the limitations of previous methods for the detection of specific exosomes (e.g., cancerous exosomes).
This study presents sensitive, specific micro-aptasensors for the detection of cancerous exosomes. Our micro-aptasensors fuse the merits of microfabricated electrodes, electrochemical analysis, hybridization chain reaction (HCR)-based signal amplification, and multiple aptamers. In our protocol, long HCR products can attach multiple avidin-horseradish peroxidases (HRPs) for efficient signal amplification for the sensitive detection of exosomes in the sample43,44,45,46. In addition, by introducing anti-EpCAM aptamers, our micro-aptasensors enable the detection of EpCAM-positive exosomes with high specificity. Based on these mechanisms, our micro-aptasensors achieve the sensitive, specific, and reliable detection of exosomes secreted by cancer cells and quantitative evaluation of the exosome concentration. Proof-of-concept experiments have been performed to detect exosomes secreted by multiple cell lines as well as exosomes isolated from patient serum samples. The performance of our micro-aptasensors was successfully validated by detecting lung cancer exosomes in serum samples from early- and late-stage lung cancer patients. We believe our developed micro-aptasensors have the potential to become a powerful exosome-based disease diagnosis platform.
Results and discussion
Mechanism of the electrochemical micro-aptasensors
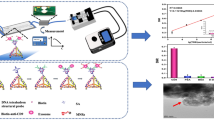
As shown in Fig. 1, our micro-aptasensor is composed of microelectrodes modified with CD63 aptamers and a microfluidic chamber. The details of the sensor fabrication and modification procedures can be found in the Experimental Section (Fig. S1). The mechanism of our electrochemical micro-aptasensor is illustrated in Fig. 1. When the HCR-exosome samples were added to the microfluidic chamber, the CD63 aptamers on the microelectrodes captured the exosomes, and the anti-EpCAM aptamers enabled HCR. Biotins labeled with H1 and H2 were used to bind with the anti-EpCAM aptamers and further produce long chains with multiple biotins. This process allows both signal amplification and the specific recognition of cancerous exosomes. As shown in Fig. 1c, a sandwich structure composed of CD63, exosomes, and anti-EpCAM-embedded HCR products was formed. For the electrochemical analysis, the current signals, which were generated through a reaction between the HRP enzyme and TMB/H2O2, were monitored by an electrochemical analyzer through microelectrodes. Details of the electrochemical analysis procedures are given in the Experimental Section. Due to the specific recognition between avidin-HRP and biotin, the current signal generated through the enzyme reaction is directly related to the amount of HRP on the HCR products and the exosome concentration.
Device optimization
To determine the optimal concentration of CD63 aptamers for the modification of our micro-aptasensors, we performed experiments with five different concentrations (0.1, 0.5, 1.0, 2.5, and 5.0 µM) of CD63. As shown in Fig. 2a, as the CD63 concentration was increased from 0.1 to 1.0 µM, the measured current increased. As the CD63 concentration was further increased from 1.0 to 5.0 µM, the current remained nearly constant. Therefore, a 1.0 µM concentration of CD63 was selected for electrode modification in this study.
a Measured current values for different CD63 concentrations that are used for the modification of microelectrodes. b Gel electrophoresis results for investigating the influence of initiator (I) concentrations on HCR. Lanes 1 and 12 are for DNA ladder markers. Lanes 2-5 are H1, H2, I, EpCAM, and a mixture of H1, H2, and EpCAM, respectively. Lanes 7-10 are for four different initiator concentrations 1.0, 0.5, 0.2, and 0.01 μM in solutions with 1 μM H1 and 1 μM H2, respectively. Lane 11 is for the mixture of I, H1, H2, and EpCAM.
To maximize the signal amplification effect with longer HCR products, we optimized the concentration of initiator aptamers for HCR according to a previous method47. As shown in Fig. 2b, when the concentration of initiator aptamers (lane 9) was 0.2 μM, the HCR product length was longer than the lengths for other concentrations. Moreover, the length of the HCR product with the anti-EpCAM aptamer (lane 11) increased slightly compared to the length of the HCR product (lane 9) without the anti-EpCAM aptamer. This result showed that the anti-EpCAM aptamer can successfully hybridize with H1, H2, and I. The longest product was over 2,000 base pairs. This showed that the HCR method is efficient for signal amplification.
Sensitivity and detection limit of the electrochemical micro-aptasensors
To characterize the sensitivity and detection limit of our micro-aptasensors, exosomes isolated from MCF-7 cell culture medium were used48. The isolated exosomes were characterized using transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). As shown in Fig. 3a, the isolated exosomes exhibited a typical cup-shaped appearance. The size distribution of exosomes ranged from 30 to 200 nm, and the concentration was 1.91×108 exosomes/mL, as shown in Fig. 3b.
The current responses to MCF-7-derived exosomes at different concentrations (1×103, 1×104, 1×105, 1×106, 2.5×106 and 1.0×107 exosomes/mL) were measured. As shown in Fig. 4a and b, as the concentrations of exosomes were increased from 2.5×103 to 2.5×106 exosomes/mL, the measured current values significantly increased. As the concentrations were further increased from 2.5×106 to 1.0×107 exosomes/mL, the current value increased only slightly because the analyte-capture capability of our micro-aptasensors gradually achieved saturation. Through a data regression step, a linear relationship (shown in Fig. 4c) was found between the current value and the logarithm of the exosome concentration, as expressed by the equation Ic = −0.08305 log c −0.8012 (R2=0.9910), where Ic represents the signal intensity, and c represents the exosome concentration. As shown in Table S1 in the Supporting Information, our aptasensor exhibits high sensitivity with a detection limit of 0.5 exosomes/μL, which is significantly lower than the limits of most published methods23,24,25,26,27,49,50,51. The high sensitivity of our method is related to the high electron transfer efficiency of our microelectrodes and the HCR amplification strategy.
Specificity and reliability of the electrochemical micro-aptasensors
To investigate the specificity of our micro-aptasensors, experiments were performed with exosomes derived from multiple cell lines, including EpCAM-positive cell lines (MCF-7 and PC3) and EpCAM-negative cell lines (Jurkat, HeLa, and U937). As shown in Fig. 5a, only exosomes derived from EpCAM-positive cell lines gave high current responses with our micro-aptasensors. For EpCAM-negative cell lines, the measured current values were much lower. These results proved that our method has high specificity for EpCAM-positive exosomes.
a Comparison among measured current values for exosomes isolated from the culture media of EpCAM-positive (MCF-7 and PC-3) and EpCAM-negative (U937, Jurkat, and HeLa) cell lines. +: EpCAM-positive; −: EpCAM-negative. b Comparison among measured current values for 5 different electrochemical micro-aptasensors operated under the same conditions.
To investigate the reliability of our method, micro-aptasensors were fabricated and used under the same conditions to detect EpCAM-positive exosomes at a concentration of 2.0×106 exosomes/mL. As shown in Fig. 5b, the current values measured from five different sensors had a small relative standard deviation of 1.86%. This relatively small standard deviation showed that our method gives consistent detection results among different devices.
Detection of cancerous exosomes isolated from human serum samples
To assess their potential in preclinical/clinical applications, the developed micro-aptasensors were used for detecting exosomes in human serum samples. Exosomes were isolated from the serum samples of 2 healthy individuals as well as 2 early-stage and 2 late-stage lung cancer patients. Figure 6a and b show the exosome detection results by NTA and our method, respectively. The exosome amounts in the early-stage and late-stage lung cancer samples were significantly higher than the exosome amounts in the healthy patient samples. The exosome amount in the late-stage lung cancer sample was significantly higher than the exosome amounts in both the early-stage lung cancer and the healthy patient samples. Compared to the NTA analysis (Fig. 6a), lower exosome concentrations were detected by our method (Fig. 6b). This difference is likely because the NTA-based method detects not only cancerous exosomes but also noncancerous exosomes51, while our method detects only cancerous exosomes. These experimental results demonstrated that our electrochemical micro-aptasensors can not only detect cancerous exosomes in patient samples and distinguish them from noncancerous exosomes but also can differentiate samples from early-stage lung cancer patients from those from late-stage patients. In this regard, our electrochemical micro-aptasensors have great potential for early cancer detection and the identification of the corresponding stage of cancer.
Conclusions
We have established sensitive, specific, and reliable electrochemical micro-aptasensors for the detection of exosomes by taking advantage of both electrochemical analysis and HCR-based signal amplification. With the introduction of anti-EpCAM aptamers, our developed micro-aptasensors can achieve high specificity for the detection of EpCAM-positive cancerous exosomes. Moreover, the microelectrodes with the HCR-based signal amplification approach greatly improve the detection sensitivity. In addition to exosomes from cell culture medium, our micro-aptasensors have been successfully applied for the detection of cancerous exosomes in serum samples of early-stage and late-stage lung cancer patients, shedding new light on early cancer diagnosis.
Experimental section
Exosome isolation
Two EpCAM-positive cell lines (MCF-7 and PC3) and three EpCAM-negative cell lines (Jurkat, HeLa, and U937) were cultured to secrete exosomes. MCF-7 and HeLa cells were cultured in DMEM (Gibco, Life Technologies, MA, USA) with 5% exosome-depleted fetal bovine serum (Gibco, Thermo Fisher, MA, USA) and 1% penicillin streptomycin (Gibco, Life Technologies, MA, USA). PC3, Jurkat, and U937 cells were cultured in RPMI-1640 (Gibco, Life Technologies, MA, USA) with 5% exosome-depleted fetal bovine serum and 1% penicillin streptomycin. To isolate exosomes from the culture medium, the medium was centrifuged at 2000×g for 10 min at room temperature. The supernatant was then centrifuged at 10,000 g for 30 min in an ultracentrifuge (Optima XE, Beckman, CA, USA) to remove large microvesicles, such as ectosomes and oncosomes. The supernatant from this centrifugation was further centrifuged at 100,000 g for 2 h. After all the centrifugation steps, the collected pellet contained only exosomes and other small particles, such as lipoproteins. The collected pellet was resuspended in phosphate-buffered saline (PBS, Invitrogen, CA, USA) for further analysis.
To isolate exosomes from the serum samples of lung cancer patients, exosome isolation reagent (Thermo Fisher, MA, USA) was used following the manufacturer’s instructions starting with 200 μL of each serum sample. All serum samples were obtained from the Department of Ophthalmology at Duke University and represented 2 healthy individuals (H), 2 early-stage lung cancer patients, and 2 late-stage of lung cancer patients (P). The samples used for this study were approved by the Duke University Health System Institutional Review Board, and all subjects gave written informed consent (Protocol ID: Pro00012914).
Fabrication and modification of micro-aptasensors
As shown in Fig. 1, the micro-aptasensor is composed of microelectrodes modified with CD63 aptamers and a microfluidic chamber. The microelectrodes were fabricated through standard lithography23, E-beam evaporation, and lift-off procedures on a 500 μm thick fused silica substrate (University wafer, MA, USA). During the E-beam evaporation process, a double-metal layer with Ti (10 nm, bottom) and Au (400 nm, top) was deposited on the substrate to form microelectrodes. After fabrication, the microelectrodes were modified. The CD63 aptamer stock solution (100 μM) was mixed with 10 mM tris(2-carboxyethyl) phosphine (TCEP, Sigma Aldrich, MO, USA) for 1 h to cleave the disulfide bonds, and then 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Sigma Aldrich, MO, USA) buffer was used to further dilute the CD63 aptamers to 1 μM. Then, 30 μL of the prepared solution of CD63 aptamers was placed on the microelectrodes for overnight incubation. After incubation, the microelectrodes were washed with deionized (DI) water. Next, 30 μL of 1 mM 6-mercapto-1-hexanol (MCH, Sigma Aldrich, MO, USA) was added to the microelectrodes for 15 min. To remove the excess MCH, the microelectrodes were washed thoroughly with DI water. In the experiments, a polydimethylsiloxane (PDMS) chamber was used to prevent lateral spreading of the liquid sample. Briefly, PDMS was first prepared through the standard mold-replication method52 and then cut to the appropriate size and drilled. Finally, the open PDMS chamber was bonded to the glass substrate to expose the patterned electrodes and prevent the added solution from leaking.
Hybridization chain reaction (HCR)
To perform HCR, 2 μM H1 and 2 μM H2 (Table 1) were prepared by diluting stock solutions with Tris buffer containing 50 mM MgCl2 (TM) buffer (20 mM, pH 8.0). Then, the prepared H1 and H2 solutions were heated to 95 °C for 10 min and immediately cooled to room temperature using ice. A solution of 10 μM anti-EpCAM aptamers and 10 μM initiator (I) strands was prepared. The above prepared solutions were then mixed to obtain the prepared hybridization mixture containing anti-EpCAM aptamers I, H1, and H2 at concentrations of 0.2, 0.2, 1, and 1 μM, respectively. The prepared hybridization mixture was kept at 37 °C overnight to obtain the HCR products.
Exosome detection through electrochemical analysis
For exosome detection, HCR-exosomes were prepared by adding approximately 1 million exosomes to 1 mL of a solution of HCR products and then shaken at 4 °C for 1 h (Fig. S2A in the Supporting Information). Next, 40 μL of HCR-exosome solution was added to the CD63-modified microelectrodes and incubated for 40 min at room temperature (Fig. S2B in the Supporting Information). Then, 2.5 μL of 10 μm/mL avidin−HRP (horseradish peroxidase coupled with avidin, Sigma Aldrich, MO, USA) was added to the microelectrodes. After 15 min at room temperature, the microelectrodes were rinsed to prepare for electrochemical analysis. An electrochemical analyzer (CHI 800B, TX, USA) was used for the analysis. 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (Neogen K-blue low-activity substrate, Shanghai, China) in H2O2 was used as the reaction solution38. The electroreduction current was measured for 50 s with an applied voltage of 100 mV after the HRP redox reaction reached steady state.
Characterization methods
Transmission electron microscopy (TEM) characterization
TEM was conducted according to the protocols outlined in the Supporting Information and used for morphological characterization of the exosomes.
Nanoparticle tracking analysis (NTA)
The size distribution and concentration of the isolated exosomes were characterized with an NTA (Nanosight LM10, Malvern, USA) system.
Electrophoresis
A 40 mL 1% agarose gel containing 4 μL of SYBR safe DNA stain (Thermo Fisher Scientific, MA, USA) was prepared with TAE buffer (40 mM Tris-acetate and 1 mM EDTA, pH 8.0). Gel electrophoresis of HCR products was performed at 100 V for 40 min using a Bio-Rad PowerPacTM Basic Electrophoresis Analyzer (Bio-Rad, CA, USA) and imaged using a Bio-Rad ChemDoc Touch Imaging System (Bio-Rad, CA, USA).
DNA sequences
All the DNA sequences used in this study were synthesized by Sangon Biotech (Shanghai, China). Their sequences are given in Table 1.
References
Simons, M. & Raposo, G. Exosomes - vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 (2009).
Théry, C., Ostrowski, M. & Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009).
Taylor, D. D. & Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 116, 153 (2010).
Saman, S. et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 287, 3842–3849 (2012).
Gupta, S. K., Bang, C. & Thum, T. Circulating MicroRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ. Cardiovasc. Genet. 3, 484–488 (2010).
Balaj, L. et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2, 1–9 (2011).
Skog, J. et al. Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Wan, Y. et al. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nat. Biomed. Eng. 1, 1–24 (2017).
Hou, B. R., Jiang, C., Wang, Z. N. & Ren, H. J. Exosome-mediated crosstalk between microglia and neural stem cells in the repair of brain injury. Neural Regen. Res. 15, 1023–1024 (2020).
Whiteside, T. L. Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 74, 103–141 (2017).
Lazar, I. et al. Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: a novel mechanism linking obesity and cancer. Cancer Res. 76, 4051–4057 (2016).
Xu, R. et al. Extracellular vesicles in cancer — implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 15, 617–638 (2018).
Luga, V. et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 (2012).
Li, K., Hong, Z. P., Li, Y. X., Li, Y. & Yang, J. L. Clinical significance of CD151 expression in non-small cell lung cancer. Chin. J. Cancer Prev. Treat. 21, 34–38 (2014).
Rahbari, M., Rahbari, N., Reissfelder, C., Weitz, J. & Kahlert, C. Exosomes: novel implications in diagnosis and treatment of gastrointestinal cancer. Langenbeck’s Arch. Surg. 401, 1097–1110 (2016).
Shao, H. et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 6, 1–9 (2015).
Zhang, Q. et al. Universal Ti 3 C 2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal. Chem. 90, 12737–12744 (2018).
He, F., Wang, J., Yin, B. C. & Ye, B. C. Quantification of exosome based on a copper-mediated signal amplification strategy. Anal. Chem. 90, 8072–8079 (2018).
Xia, Y. et al. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 92, 8–15 (2017).
Xia, Y. et al. A nature-inspired colorimetric and fluorescent dual-modal biosensor for exosomes detection. Talanta 214, 120851 (2020).
Zhao, L., Sun, R., He, P. & Zhang, X. Ultrasensitive detection of exosomes by target triggered 3D DNA walking machine and exonuclease III-assisted electrochemical ratiometric biosensing. Anal. Chem. 91, 14773–14779 (2019).
Zhou, Q. et al. Development of an aptasensor for electrochemical detection of exosomes. Methods 97, 88–93 (2016).
Huang, R. et al. A sensitive aptasensor based on a Hemin/G-quadruplex-assisted signal amplification strategy for electrochemical detection of gastric cancer exosomes. Small 15, 1–7 (2019).
Dong, H. et al. Highly sensitive electrochemical detection of tumor exosomes based on aptamer recognition-induced multi-DNA release and cyclic enzymatic amplification. Anal. Chem. 90, 4507–4513 (2018).
Cao, Y. et al. A catalytic molecule machine-driven biosensing method for amplified electrochemical detection of exosomes. Biosens. Bioelectron. 141, 111397 (2019).
An, Y., Jin, T., Zhu, Y., Zhang, F. & He, P. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens. Bioelectron. 142, 111503 (2019).
Stremersch, S. et al. Identification of individual exosome-like vesicles by surface enhanced raman spectroscopy. Small 12, 3292–3301 (2016).
Ning, C. F., Wang, L., Tian, Y. F., Yin, B. C. & Ye, B. C. Multiple and sensitive SERS detection of cancer-related exosomes based on gold-silver bimetallic nanotrepangs. Analyst 145, 2795–2804 (2020).
Zhu, L. et al. Label-free quantitative detection of tumor-derived exosomes through surface plasmon resonance imaging. Anal. Chem. 86, 8857–8864 (2014).
Wang, L. et al. Bridging exosome and liposome through zirconium-phosphate coordination chemistry: a new method for exosome detection. Chem. Commun. 55, 2708–2711 (2019).
Zeng, R. et al. Nano energy single-atom platinum nanocatalyst-improved catalytic efficiency with enzyme-DNA supermolecular architectures. Nano Energy 74, 104931 (2020).
Luo, X., An, M., Cuneo, K. C., Lubman, D. M. & Li, L. High-performance chemical isotope labeling liquid chromatography mass spectrometry for exosome metabolomics. Anal. Chem. 90, 8314–8319 (2018).
Zhu, C., Li, L., Wang, Z., Irfan, M. & Qu, F. Recent advances of aptasensors for exosomes detection. Biosens. Bioelectron. 160, 112213 (2020).
Fu, Y., Jiang, C., Tofaris, G. K. & Davis, J. J. A facile impedimetric analysis of neuronal exosome markers in Parkinson’s disease diagnostics. Anal. Chem. 92, 13647–13651 (2020).
Doldán, X., Fagúndez, P., Cayota, A., Laíz, J. & Tosar, J. P. Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal. Chem. 88, 10466–10473 (2016).
Jeong, S. et al. Integrated magneto-electrochemical sensor for exosome analysis. ACS Nano 10, 1802–1809 (2016).
Wen, J. T., Bohorquez, K. & Tsutsui, H. Polydiacetylene-coated polyvinylidene fluoride strip aptasensor for colorimetric detection of zinc(II). Sens. Actuators B Chem. 232, 313–317 (2016).
Chen, C. et al. Development of a structure-switching aptamer-based nanosensor for salicylic acid detection. Biosens. Bioelectron. 140, 111342 (2019).
Ding, X., Mauk, M. G., Yin, K., Kadimisetty, K. & Liu, C. Interfacing pathogen detection with smartphones for point-of-care applications. Anal. Chem. 91, 655–672 (2019).
Wu, Y. et al. Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano 11, 3943–3949 (2017).
Fu, Z., Lu, Y.-C. & Lai, J. J. Recent advances in biosensors for nucleic acid and exosome detection. Chonnam Med. J. 55, 86 (2019).
Zhang, K., Lv, S., Zhou, Q. & Tang, D. CoOOH nanosheets-coated g-C3N4/CuInS2 nanohybrids for photoelectrochemical biosensor of carcinoembryonic antigen coupling hybridization chain reaction with etching reaction. Sens. Actuators B Chem. 307, 127631 (2020).
Zhang, B. et al. DNA-based hybridization chain reaction for amplified bioelectronic signal and ultrasensitive detection of proteins. Anal. Chem. 84, 5392–5399 (2012).
Zeng, R. et al. Photoelectrochemical bioanalysis of antibiotics on rGO-Bi 2 WO 6 -Au based on branched hybridization chain reaction. Biosens. Bioelectron. 133, 100–106 (2019).
Gao, Z., Qiu, Z., Lu, M., Shu, J. & Tang, D. Hybridization chain reaction-based colorimetric aptasensor of adenosine 5′-triphosphate on unmodified gold nanoparticles and two label-free hairpin probes. Biosens. Bioelectron. 89, 1006–1012 (2017).
Dirks, R. M. & Pierce, N. A. Triggered amplification by hybridization chain reaction. Proc. Natl Acad. Sci. USA 101, 15275–15278 (2004).
Huang, Q. et al. Gelatin nanoparticle-coated silicon beads for density-selective capture and release of heterogeneous circulating tumor cells with high purity. Theranostics 8, 1624–1635 (2018).
Yin, X., Hou, T., Huang, B., Yang, L. & Li, F. Aptamer recognition-trigged label-free homogeneous electrochemical strategy for an ultrasensitive cancer-derived exosome assay. Chem. Commun. 55, 13705–13708 (2019).
Zhao, L., Sun, R., He, P. & Zhang, X. Ultrasensitive detection of exosomes by target-triggered three-dimensional DNA walking machine and exonuclease III-assisted electrochemical ratiometric biosensing. Anal. Chem. 91, 14773–14779 (2019).
Sun, Y., Jin, H., Jiang, X. & Gui, R. Assembly of black phosphorus nanosheets and MOF to form functional hybrid thin-film for precise protein capture, dual-signal and intrinsic self-calibration sensing of specific cancer-derived exosomes. Anal. Chem. 92, 2866–2875 (2020).
Sun, C. et al. Thin film Gallium nitride (GaN) based acoustofluidic tweezer: modelling and microparticle manipulation. Ultrasonics 108, 1–7 (2020).
Acknowledgements
We acknowledge the support of National Institutes of Health (NIH) grants U18TR003778 and R01GM135486. Wenfen Zhang acknowledges the support of the National Natural Science Foundation of China (22004109) and the China Scholarship Council. Guozhen Liu acknowledges the support of the Chinese University of Hong Kong, Shenzhen providing the University Development fund and Presidential Fellow fund. Joseph Rich acknowledges that this material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1644868.
Author information
Authors and Affiliations
Contributions
W.Z. and T.J.H. designed this research. W.Z. fabricated the devices, conducted the exosome experiments, and prepared the manuscript. Z.T., G.L., J.R., A.S., and Q.M. analyzed the data and edited the manuscript. S.Y. fabricated all the electrodes used in the experiments. M.K. conducted exosome isolation. S.G.Z. and P.H. analyzed the data. T.J.H., E.P., and S.S.Z. supervised the work and edited the manuscript. Z.L. drew the schematic figure. All authors have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
T.J.H. has cofounded a start-up company, Ascent Bio-Nano Technologies Inc., to commercialize technologies involving acoustofluidics and acoustic tweezers. The remaining authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, W., Tian, Z., Yang, S. et al. Electrochemical micro-aptasensors for exosome detection based on hybridization chain reaction amplification. Microsyst Nanoeng 7, 63 (2021). https://doi.org/10.1038/s41378-021-00293-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41378-021-00293-8
This article is cited by
-
Advanced extracellular vesicle bioinformatic nanomaterials: from enrichment, decoding to clinical diagnostics
Journal of Nanobiotechnology (2023)
-
Fluorescent aptasensor based on the MNPs-CRISPR/Cas12a-TdT for the determination of nasopharyngeal carcinoma-derived exosomes
Microchimica Acta (2023)
-
Current status and outlook of advances in exosome isolation
Analytical and Bioanalytical Chemistry (2022)