Abstract
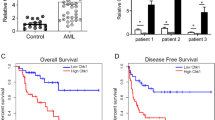
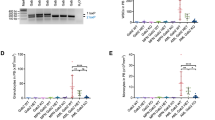
Internal tandem duplication (ITD) mutations within the FMS-like tyrosine kinase-3 (FLT3) occur in up to 25% of acute myeloid leukemia (AML) patients and indicate a very poor prognosis. The role of long noncoding RNAs (lncRNAs) in FLT3-ITD AML progression remains unexplored. We identified a novel lncRNA, SNHG29, whose expression is specifically regulated by the FLT3-STAT5 signaling pathway and is abnormally down-regulated in FLT3-ITD AML cell lines. SNHG29 functions as a tumor suppressor, significantly inhibiting FLT3-ITD AML cell proliferation and decreasing sensitivity to cytarabine in vitro and in vivo models. Mechanistically, we demonstrated that SNHG29’s molecular mechanism is EP300-binding dependent and identified the EP300-interacting region of SNHG29. SNHG29 modulates genome-wide EP300 genomic binding, affecting EP300-mediated histone modification and consequently influencing the expression of varies downstream AML-associated genes. Our study uncovers a novel molecular mechanism for SNHG29 in mediating FLT3-ITD AML biological behaviors through epigenetic modification, suggesting that SNHG29 could be a potential therapeutic target for FLT3-ITD AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52.
Kiyoi H, Kawashima N, Ishikawa Y. FLT3 mutations in acute myeloid leukemia: Therapeutic paradigm beyond inhibitor development. Cancer Sci. 2020;111:312–22.
Takahashi S. Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications. J Hematol Oncol. 2011;4:13.
Kazi JU, Rönnstrand L. FMS-like tyrosine kinase 3/FLT3: from basic science to clinical implications. Physiol Rev. 2019;99:1433–66.
Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med. 2019;381:1728–40.
McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJD, Eastburn DJ, et al. Clonal selection with RAS pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer Discov. 2019;9:1050–63.
Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. 2013;70:4785–94.
Delás MJ, Sabin LR, Dolzhenko E, Knott SR, Munera Maravilla E, Jackson BT, et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. Elife. 2017;6:e25607.
Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. 2014;5:5220.
Sun LY, Li XJ, Sun YM, Huang W, Fang K, Han C, et al. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1. Mol Cancer. 2018;17:127.
Feng Y, Hu S, Li L, Zhang S, Liu J, Xu X, et al. LncRNA NR-104098 Inhibits AML Proliferation and Induces Differentiation Through Repressing EZH2 Transcription by Interacting With E2F1. Front Cell Dev Biol. 2020;8:142.
Papaioannou D, Petri A, Dovey OM, Terreri S, Wang E, Collins FA, et al. The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat Commun. 2019;10:5351.
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y, et al. HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell. 2019;36:645–59.e8.
Lin N, Yao Z, Xu M, Chen J, Lu Y, Yuan L, et al. Long noncoding RNA MALAT1 potentiates growth and inhibits senescence by antagonizing ABI3BP in gallbladder cancer cells. J Exp Clin Cancer Res. 2019;38:244.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Franco HL, Nagari A, Malladi VS, Li W, Xi Y, Richardson D, et al. Enhancer transcription reveals subtype-specific gene expression programs controlling breast cancer pathogenesis. Genome Res. 2018;28:159–70.
Cai T, Zhang Q, Wu B, Wang J, Li N, Zhang T, et al. LncRNA-encoded microproteins: a new form of cargo in cell culture-derived and circulating extracellular vesicles. J Extracell Vesicles. 2021;10:e12123.
Sun L, Wang W, Han C, Huang W, Sun Y, Fang K, et al. The oncomicropeptide APPLE promotes hematopoietic malignancy by enhancing translation initiation. Mol Cell. 2021;81:4493–508.e9.
Li Y, Liu X, Cui X, Tan Y, Wang Q, Wang Y, et al. LncRNA PRADX-mediated recruitment of PRC2/DDX5 complex suppresses UBXN1 expression and activates NF-κB activity, promoting tumorigenesis. Theranostics. 2021;11:4516–30.
Wang FW, Cao CH, Han K, Zhao YX, Cai MY, Xiang ZC, et al. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J Clin Invest. 2019;129:727–43.
Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–31.
Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003;9:2140–50.
Uttarkar S, Dassé E, Coulibaly A, Steinmann S, Jakobs A, Schomburg C, et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood. 2016;127:1173–82.
He P, Zhang C, Ji Y, Ge MK, Yu Y, Zhang N, et al. Epithelial cells-enriched lncRNA SNHG8 regulates chromatin condensation by binding to Histone H1s. Cell Death Differ. 2022;29:1569–81.
Liu HT, Zou YX, Zhu WJ, Sen L, Zhang GH, Ma RR, et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2022;29:627–41.
Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol. 2019;55:106–19.
Buquicchio FA, Satpathy AT. Interrogating immune cells and cancer with CRISPR-Cas9. Trends Immunol. 2021;42:432–46.
Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–5.
Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045.
Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 2021;41:109–20.
Yusenko MV, Biyanee A, Andersson MK, Radetzki S, von Kries JP, Stenman G, et al. Proteasome inhibitors suppress MYB oncogenic activity in a p300-dependent manner. Cancer Lett. 2021;520:132–42.
Saeed S, Logie C, Francoijs KJ, Frigè G, Romanenghi M, Nielsen FG, et al. Chromatin accessibility, p300, and histone acetylation define PML-RARα and AML1-ETO binding sites in acute myeloid leukemia. Blood. 2012;120:3058–68.
Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–76.
He ZX, Wei BF, Zhang X, Gong YP, Ma LY, Zhao W. Current development of CBP/p300 inhibitors in the last decade. Eur J Med Chem. 2021;209:112861.
Huang YH, Cai K, Xu PP, Wang L, Huang CX, Fang Y, et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct Target Ther. 2021;6:10.
Garcia-Carpizo V, Ruiz-Llorente S, Sarmentero J, González-Corpas A, Barrero MJ. CREBBP/EP300 bromodomain inhibition affects the proliferation of AR-positive breast cancer cell lines. Mol Cancer Res. 2019;17:720–30.
Palazzo AF, Koonin EV. Functional long non-coding RNAs evolve from junk transcripts. Cell. 2020;183:1151–61.
Rack JGM, Lutter T, Kjæreng Bjerga GE, Guder C, Ehrhardt C, Värv S, et al. The PHD finger of p300 influences its ability to acetylate histone and non-histone targets. J Mol Biol. 2014;426:3960–72.
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95.
Funding
This work was supported by the Fujian Provincial Health and Family Planning Commission Medical Innovation Fund (No.: 2018-CX-5 and 2019-CX-2), the Natural Science Foundation of Fujian Province, Fujian, China (No.: 2021J01389, 2020J011098, and 2022J0112), and the High-level Hospital Foster Grants from Fujian Provincial Hospital, Fujian Province, China (No.: 2019HSJJ15).
Author information
Authors and Affiliations
Contributions
SJ and LS conceptualized the study; SJ, LS, ZJ, YXL, and CDN designed the experiments; LS, ZJ, LYB, and CZZ performed the experiments; SJ, LS, LYB, and ZJ produced the figures; LS and SJ wrote the original manuscript; SJ and CWM reviewed and edited the manuscript; SJ, LS, and CBY were responsible for funding application; SJ provided supervision. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, S., Zhou, J., Ye, X. et al. A novel lncRNA SNHG29 regulates EP300- related histone acetylation modification and inhibits FLT3-ITD AML development. Leukemia 37, 1421–1434 (2023). https://doi.org/10.1038/s41375-023-01923-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01923-y