Abstract
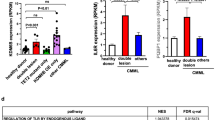
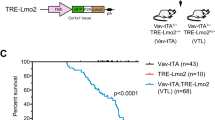
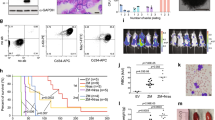
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematopoietic neoplasm resulting from the malignant transformation of T-cell progenitors. While activating NOTCH1 mutations are the dominant genetic drivers of T-ALL, epigenetic dysfunction plays a central role in the pathology of T-ALL and can provide alternative mechanisms to oncogenesis in lieu of or in combination with genetic mutations. The histone demethylase enzyme KDM6A (UTX) is also recurrently mutated in T-ALL patients and functions as a tumor suppressor. However, its gene paralog, KDM6B (JMJD3), is never mutated and can be significantly overexpressed, suggesting it may be necessary for sustaining the disease. Here, we used mouse and human T-ALL models to show that KDM6B is required for T-ALL development and maintenance. Using NOTCH1 gain-of-function retroviral models, mouse cells genetically deficient for Kdm6b were unable to propagate T-ALL. Inactivating KDM6B in human T-ALL patient cells by CRISPR/Cas9 showed KDM6B-targeted cells were significantly outcompeted over time. The dependence of T-ALL cells on KDM6B was proportional to the oncogenic strength of NOTCH1 mutation, with KDM6B required to prevent stress-induced apoptosis from strong NOTCH1 signaling. These studies identify a crucial role for KDM6B in sustaining NOTCH1-driven T-ALL and implicate KDM6B as a novel therapeutic target in these patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Primary RNA-sequencing data are available through NCBI Gene Expression Omnibus under GEO accession number GSE214576.
References
Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–43.
Uckun FM, Gaynon PS, Sensel MG, Nachman J, Trigg ME, Steinherz PG, et al. Clinical features and treatment outcome of childhood T-lineage acute lymphoblastic leukemia according to the apparent maturational stage of T-lineage leukemic blasts: a Children’s Cancer Group study. J Clin Oncol. 1997;15:2214–21.
Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21:3616–22.
Oudot C, Auclerc MF, Levy V, Porcher R, Piguet C, Perel Y, et al. Prognostic factors for leukemic induction failure in children with acute lymphoblastic leukemia and outcome after salvage therapy: the FRALLE 93 study. J Clin Oncol. 2008;26:1496–503.
Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129:1113–23.
Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16:494–507.
Ferrando AA The role of NOTCH1 signaling in T-ALL. Hematology Am Soc Hematol Educ Program. 2009:353–61.
Huether R, Dong L, Chen X, Wu G, Parker M, Wei L, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630.
Neumann M, Vosberg S, Schlee C, Heesch S, Schwartz S, Gokbuget N, et al. Mutational spectrum of adult T-ALL. Oncotarget. 2015;6:2754–66.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63.
Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301.
Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–17.
Arcipowski KM, Martinez CA, Ntziachristos P. Histone demethylases in physiology and cancer: a tale of two enzymes, JMJD3 and UTX. Curr Opin Genet Dev. 2016;36:59–67.
Wei Y, Zheng H, Bao N, Jiang S, Bueso-Ramos CE, Khoury J, et al. KDM6B overexpression activates innate immune signaling and impairs hematopoiesis in mice. Blood Adv. 2018;2:2491–504.
Ohguchi H, Harada T, Sagawa M, Kikuchi S, Tai YT, Richardson PG, et al. KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia. 2017;31:2661–9.
Mallaney C, Ostrander EL, Celik H, Kramer AC, Martens A, Kothari A, et al. Kdm6b regulates context-dependent hematopoietic stem cell self-renewal and leukemogenesis. Leukemia. 2019;33:2506–21.
Iwamori N, Iwamori T, Matzuk MM. H3K27 demethylase, JMJD3, regulates fragmentation of spermatogonial cysts. PLoS ONE. 2013;8:e72689.
Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–6.
Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–9.
Nakka K, Hachmer S, Mokhtari Z, Kovac R, Bandukwala H, Bernard C, et al. JMJD3 activated hyaluronan synthesis drives muscle regeneration in an inflammatory environment. Science. 2022;377:666–9.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Srivastava S, Sahu U, Zhou Y, Hogan AK, Sathyan KM, Bodner J, et al. NOTCH1-driven UBR7 stimulates nucleotide biosynthesis to promote T cell acute lymphoblastic leukemia. Sci Adv. 2021;7:eabc9781.
Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–6.
Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–91.
Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–7.
Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–44.
Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605.
Salminen A, Kaarniranta K, Hiltunen M, Kauppinen A. Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. J Mol Med. 2014;92:1035–43.
Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX, and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007;449:731–4.
Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–7.
Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, et al. Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep. 2016;17:1453–61.
Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective Jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012;488:404–8.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004;306:269–71.
Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–51.
Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–35.
O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–24.
Kourtis N, Lazaris C, Hockemeyer K, Balandran JC, Jimenez AR, Mullenders J, et al. Oncogenic hijacking of the stress response machinery in T cell acute lymphoblastic leukemia. Nat Med. 2018;24:1157–66.
Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood 2009;113:1689–98.
Wendorff AA, Quinn SA, Rashkovan M, Madubata CJ, Ambesi-Impiombato A, Litzow MR, et al. Phf6 loss enhances HSC self-renewal driving tumor initiation and leukemia stem cell activity in T-ALL. Cancer Discov. 2019;9:436–51.
Figueroa ME, Chen SC, Andersson AK, Phillips LA, Li Y, Sotzen J, et al. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J Clin Invest. 2013;123:3099–111.
Mullighan CG, Downing JR. Global genomic characterization of acute lymphoblastic leukemia. Semin Hematol. 2009;46:3–15.
Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–8.
Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 2015;125:13–21.
Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–406.
Acknowledgements
We thank all members of the Challen laboratory for ongoing contributions and critical discussion. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and the Institute of Clinical and Translational Sciences (ICTS) at Washington for the use of the Tissue Procurement Core, which provided T-ALL patient samples. Support for the procurement of human samples was provided by P50CA171963. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant P30CA091842 and the ICTS is funded by the National Institutes of Health’s NCATS Clinical and Translational Science Award (CTSA) program grant UL1TR002345. We thank the Roswell Park Hematological Procurement Shared Resource for T-ALL specimens which is supported by the NCI Cancer Center Support Grant 5P30CA001656. We thank the Siteman Cancer Center Flow Cytometry core, which is supported by NIH Cancer Center Support Grant P30CA091842. We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for genomic analyses. The Center is partially supported by NCI Cancer Center Support Grant P30CA91842. This publication is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. This work was supported by the National Institutes of Health (HL147978, CA236819, and DK124883 to G.A.C), Gabrielle’s Angel Foundation (to G.A.C), Alex’s Lemonade Stand Foundation (to G.A.C) and the Children’s Discovery Institute (to G.A.C). C.R.Z. was supported by an American Society of Hematology post-doctoral scholar award and the Edward P. Evans Center for MDS Post-Doctoral Fellowship. Z.R. and K.G. were supported by the Intramural Research Program of NIDDK, NIH. E.S.W. is supported by the Jacquie Hirsch Leukemia Research Fund. G.A.C. is a scholar of the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Contributions
Conceptualization and study design—GAC. Experimentation and data acquisition—NI, HB, AK, WMBD, TMP, CRZ, WH, GAC Data analysis—NI, ERW, ALY, DHS, GAC. Resources, and reagents—ZR, KG, ESW, APW, AC. Funding acquisition—GAC. Project administration and supervision—GAC. Manuscript preparation—GAC.
Corresponding author
Ethics declarations
Competing interests
G.A.C. has performed consulting and received research support from Incyte (unrelated to this work). The remaining authors have declared that no conflict of interest exists.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Issa, N., Bjeije, H., Wilson, E.R. et al. KDM6B protects T-ALL cells from NOTCH1-induced oncogenic stress. Leukemia 37, 728–740 (2023). https://doi.org/10.1038/s41375-023-01853-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01853-9