Abstract
The transcription factor NFE2 is overexpressed in most patients with myeloproliferative neoplasms (MPN). Moreover, mutations in NFE2, found in a subset of MPN patients, strongly predispose for transformation to acute leukemia. Transgenic mice overexpressing NFE2 as well as mice harboring NFE2 mutations display an MPN phenotype and spontaneously develop leukemia. However, the molecular mechanisms effecting NFE2-driven leukemic transformation remain incompletely understood. Here we show that the pro-leukemic histone demethylase JMJD2C constitutes a novel NFE2 target gene. JMJD2C expression is elevated in MPN patients as well as in NFE2 transgenic mice. Moreover, we show that loss of JMJD2C selectively impairs proliferation of JAK2V617F mutated cells. Our data suggest that JMJD2C represents a promising drug target in MPN and provide a rationale for further investigation in preclinical and clinical settings.
Similar content being viewed by others
Introduction
Despite discovery of the JAK2, CALR and MPL driver mutations, the molecular etiology of Myeloproliferative Neoplasms (MPN) remains incompletely understood. Current pharmacological therapies lack disease-modifying activity, necessitating the development of improved therapeutic strategies. The transcription factor NFE2 has been identified as an important player in MPN pathophysiology. A large majority of MPN patients display increased NFE2 levels [1] and transgenic NFE2 overexpressing mice develop an MPN phenotype with spontaneous transformation to acute leukemia [2]. Moreover, NFE2 mutations found in a subset of MPN patients strongly predispose for leukemic transformation both in patients and in mice [3,4,5]. As both the molecular mechanisms causing NFE2 dysregulation in MPN as well as its functional consequences are incompletely understood, their investigation may delineate novel targetable pathways in MPN.
In order to elucidate downstream effectors of NFE2 activity, we analyzed the transcriptome of human CD34+ hematopoietic stem and progenitor cells (HSPCs) following overexpression or silencing of NFE2 [6]. Subsequently, we combined our gene expression data with published NFE2 chromatin immunoprecipitation sequencing (ChIP-seq) data [7], generating a list of putative NFE2 target genes [8]. Among these potential targets, we have previously confirmed NFE2-driven transcriptional regulation of the proinflammatory cytokine interleukin 8 (IL8) and the histone demethylase JMJD1C [6, 8]. IL8 levels constitute an independent prognostic factor for survival in primary myelofibrosis (PMF) patients [9], while the pro-leukemic JMJD1C represents a promising drug target in myeloid malignancies [10]. However, we have recently shown that JMJD1C is dispensable for JAK2V617F-driven myeloproliferation [11]. Nonetheless, other NFE2 target genes remain putative novel drug candidates in MPN.
In addition to IL8 and JMJD1C, we identified the histone demethylase JMJD2C / KDM4C as a potential NFE2 target gene [6,7,8]. JMJD2C is upregulated in primary acute myeloid leukemia (AML) cells as well as in AML cell lines [10]. JMJD2C depletion in MLL-rearranged leukemia cells increases differentiation, promotes apoptosis and, upon transplantation, prolongs survival of recipient mice [12]. Ernst et al. recently showed that JMJD2C mRNA expression is also elevated in MPN patients. JMJD2C deletion attenuated growth of JAK2V617F-positive HEL cells and, upon transplantation into a xenograft model, increased survival of recipient mice [13]. These data suggest that JMJD2C constitutes an attractive novel therapeutic target in myeloid malignancies. Here we show that NFE2 directly regulates JMJD2C expression and provide mechanistic insights into the effects of altering JMJD2C activity.
Materials and methods
Materials and methods are described in the Supplementary Material.
Results
We have previously identified a list of 60 epigenetic regulators constituting potential NFE2 target genes by interrogating NFE2 ChIP-seq data in K562 cells [7]. Amongst these, the histone demethylase JMJD2C ranked highly and constitutes an attractive target due to its pro-leukemic role in myeloid malignancies [10]. We therefore investigated whether JMJD2C plays a role in the pathophysiology of Myeloproliferative Neoplasms (MPN).
The histone demethylase JMJD2C is a novel NFE2 target gene
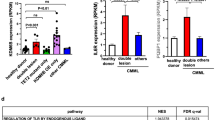
To confirm the NFE2 binding site identified by ChIP-seq at +260 bp of the JMJD2C locus ([7], Fig. 1A), we performed ChIP assays in HEL cells. A strong enrichment of NFE2 was observed around the site identified at the JMJD2C locus (Fig. 1A). Concomitantly, Jmjd2c mRNA expression was significantly elevated in the bone marrow of transgenic mice overexpressing hNFE2 compared to wild-type littermate controls (Fig. 1B). Moreover, lentiviral introduction of hNFE2 into CB3 cells, an erythroid cell line devoid of NFE2 due to viral insertion, was sufficient to induce Jmjd2c expression (Fig. 1C). Taken together, these data show that the JMJD2C locus is bound by NFE2 and that presence of NFE2 increases JMJD2C expression.
A Top: NFE2 ChIP-seq at the JMJD2C locus [7]. Bottom: ChIP PCR of HEL cells using an antibody against NFE2 or an IgG control, as depicted. PCR primers flank the predicted binding site at +260 bp or negative control sites at +4.6 kb of the JMJD2C locus and the myogenin locus. B Jmjd2c mRNA expression in BM of hNFE2tg mice (n = 12) or littermate controls (n = 9) determined by RT-qPCR. Expression is normalized to B2m expression. C Jmjd2c mRNA levels in CB3 cells following infection with pLeGO-iG-NFE2-wt or an empty control vector. GFP positive cells were sorted and Jmjd2c expression measured as described in panel 1B. D Illustration of the JMJD2C reporter gene construct. A circle indicates the predicted NFE2 binding site +260 bp relative to the transcription start site (TSS). E Cotransfection of HEK-293T cells with the JMJD2C luciferase reporter gene construct and different combinations of expression plasmids encoding NFE2 and MafG cDNAs, as depicted. F Disruption of the potential NFE2 binding site by site directed mutagenesis (crossed circle). Altered bases are shown in bold. E, F Data were normalized to the wt JMJD2C reporter gene construct cotransfected with MafG alone, which was set as one. G Exemplary western blots depicting JMJD2C expression in lysates from PB granulocytes. GAPDH was used as a loading control. H Densitometric analysis of JMJD2C western blots, n = 26 MPN patients and n = 9 healthy controls. I JMJD2C protein expression across different MPN subtypes (ET = 5, PV = 12, PMF = 9). J JMJD2C protein expression according to mutational status (JAK2V617F = 19, CALR = 7). C, E, F Data are represented as mean of at least three independent experiments. B, C, E, F, H–J *p < 0.05, **p < 0.01 by Student’s t test.
To evaluate whether NFE2 is sufficient to directly transactivate the JMJD2C promoter, we performed luciferase assays. The region of interest, bp −2220 to +522 of the JMJD2C locus, was cloned into a luciferase reporter vector (Fig. 1D) and cotransfected into HEK-293T cells together with plasmids expressing the NFE2 cDNA either with or without its binding partner MafG (Fig. 1E). As hypothesized, we observed a significant increase of luciferase activity only in the presence of both NFE2 and MafG. In turn, site directed mutagenesis of the NFE2 recognition sequence led to a significant decrease of signal intensity (Fig. 1F). NFE2 therefore binds the JMJD2C locus and mediates its transcriptional activation.
JMJD2C expression is increased in MPN patients
Since NFE2 is overexpressed in the vast majority of MPN patients [1], we investigated whether increased NFE2 in MPN patients increases JMJD2C protein expression. Peripheral blood granulocytes from MPN patients and healthy controls were analyzed by Western Blots (Fig. 1G–J). Quantification of JMJD2C protein expression from n = 26 MPN patients and n = 9 healthy controls (HC) revealed significantly elevated JMJD2C protein levels in patients with essential thrombocythemia, polycythemia vera and primary myelofibrosis (Fig. 1H, I). Likewise, JMJD2C mRNA levels were significantly elevated in post-MPN AML patients (Supplementary Fig. S1). No significant difference was observed between JAK2V617F- and CALR-mutated patients (Fig. 1J). Moreover, mRNA expression of NFE2 and JMJD2C correlated significantly (Supplementary Fig. S2), underscoring a possible role for this histone demethylase in the pathophysiology of MPN.
JMJD2C is required for the survival of JAK2V617F mutated cells
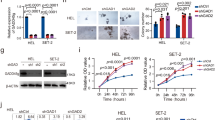
Since JMJD2C deficiency has been shown to attenuate leukemogenicity in AML (reviewed in Staehle et al. 2021), we hypothesized that JMJD2C selectively impacts proliferation of JAK2V617F cells. Hence, we determined the effect of normalizing elevated JMJD2C levels in JAK2V617F-positive cells using RNA interference. The most effective shRNA, construct #3 (Fig. 2A) was selected in UKE1 cells, yielding approximately 80–90% reduction in JMJD2C expression, as determined by both RT-qPCR and western blotting (Fig. 2B, C). JMJD2C depletion strongly increased global levels of its target histone marks H3K9me3, H3K27me3 and H3K36me3 (Fig. 2D–F).
A UKE1 cells were lentivirally transduced with vectors carrying shRNAs against JMJD2C (sh-2C #1–3) or a scrambled control (scr). Cells were harvested on day 5 for RNA isolation and subsequent analysis by RT-qPCR. B, C Validation of sh-2C #3 in UKE1, SET2 and HEL cells by RT-qPCR (B) and western blotting (C) using an anti-JMJD2C antibody, actin served as a loading control. D–F Western blotting of JMJD2C-depleted or wt UKE1, SET2 and HEL cells with antibodies against H3K9me3 (D), H3K27me3 (E) and H3K36me3 (F). Histone H3 served as a loading control. G–I Proliferation of UKE1 (G), SET2 (H) and HEL (I) cells after introduction of JMJD2C-shRNA (sh-2C) or the scrambled (scr) control. For each condition, 0.25 million cells were seeded and infected on day 1. Cells were counted every second day and used for final analysis on day 9. J–L Detection of apoptotic (AnnexinV+/PI−) and necrotic (AnnexinV+/PI+) cells on day 9 by FACS analysis in UKE1 (J), SET2 (K) and HEL (L) cells. M–O Cell cycle analysis: detection of cells in G1 (DAPI low), S (DAPI medium) and G2/M (DAPI high) phase on day 9 by FACS analysis in UKE1 (M), SET2 (N) and HEL (O) cells. P–R Competitive growth of shRNA (sh-2C or scr) infected UKE1 (P), SET2 (Q) and HEL (R) cells. 0.1 million infected (GFP+) cells were seeded at a 1:1 ratio with uninfected cells on day 1. The proportion of GFP+ cells was determined daily. G–R Data are represented as mean +/− SEM of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test.
JAK2V617F-positive UKE1, SET2 and HEL cells were lentivirally transduced with the JMJD2C shRNA or a scrambled control. Transduction efficiencies, determined by FACS analysis on day 5, reached nearly 100 % (Fig. S3). JMJD2C knockdown significantly reduced proliferation of UKE1, SET2 and HEL cells after 7–9 days (Fig. 2G–I). Moreover, Annexin V/PI staining showed an increased proportion of apoptotic and necrotic cells in JMJD2C depleted cells (Fig. 2J–L, Supplementary Fig. S4). Concomitantly, cell cycle analysis revealed reduced proportions of G2/M and increased proportions of G1 phase cells (Fig. 2M–O, Supplementary Fig. S5). In a competitive proliferation assay, we mixed JMJD2C shRNA depleted or scrambled control infected cells with uninfected cells at a 1:1 ratio. While scrambled control infected cells showed no or only a slight reduction in proliferation compared to uninfected cells, the proportion of JMJD2C depleted cells diminished rapidly, indicating that decreased JMJD2C levels confer a proliferative disadvantage (Fig. 2P–R). In contrast, in JAK2V617F-negative K562 and U937 cells, JMJD2C depletion did not affect proliferation, apoptosis or cell cycle progression and yielded no competitive disadvantage (Supplementary Fig. S6). Loss of JMJD2C expression therefore selectively impairs proliferation of JAK2V617F mutated cells, at least in part through increased apoptosis and cell cycle arrest.
Discussion
Elevated NFE2 expression and mutations in NFE2 strongly predispose for transformation to acute leukemia both in MPN patients and in murine models [2,3,4,5]. However, NFE2 itself constitutes a difficult drug target, as pharmacological modulation of transcription factors remains challenging. Thus, defining the pathways affected by elevated NFE2 levels may identify more eligible targets for MPN therapy.
Here, we demonstrate that the histone demethylase JMJD2C constitutes a novel NFE2 target gene (Fig. 1). JMJD2C plays a pro-proliferative role in selected myeloid malignancies, however, its activity is context dependent. Loss of JMJD2C does not affect BCR-ABL, AML-ETO and PML-RARA transformed cells (Supplementary Fig. S6, [12]). Conversely, in MLL-driven AML and in JAK2V617F-positive MPN, targeting JMJD2C reduces leukemogenicity by increasing differentiation, promoting cell cycle arrest and enhancing apoptosis [10, 12, 13].
As a histone demethylase, JMJD2C affects cell proliferation in myeloid malignancies via epigenetic mechanisms, removing repressive H3K9me3, H3K27me3 and activating H3K36me3 histone marks (Fig. 2D–F, [12, 13]). In MLL-driven AML, elevated JMJD2C levels increase expression of pro-proliferative targets, among them Myc, Hoxa9, and Meis1 [12], while increased JMJD2C activity in JAK2V617F-positive HEL cells prevents cellular senescence [13]. JMJD2C depletion thus affects distinct neoplastic pathways in specific clinical entities. In contrast, reduction of JMJD2C levels does not impair healthy hematopoiesis [14], arguing for a therapeutic window in MPN and AML.
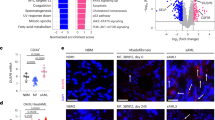
To identify changes in gene expression caused by JMJD2C depletion selectively in JAK2V617F cells, we compared RNA-seq data from two cellular models, following JMJD2C targeting: HEL (JAK2V617F) and MLL-GAS7 (JAK2 wt) (Supplementary Fig. S7). Genes regulated by HIF1A, JUN and SP1 are selectively enhanced by JMJD2C depletion in JAK2V617 positive cells, while Myc signaling and TP53 targets are altered both in JAK2V617F and JAK2 wt cells (Supplementary Fig. S7).
Currently available small molecule JMJD2C inhibitors target the enzyme’s catalytic or reader domain [15]. TACH101, an orally available, reversible, competitive KDM4-family inhibitor, recently entered a phase I/II trial for advanced and metastatic GI tumors (NCT05076552). Safety and tolerability data from this first-in-human trial will determine whether JMJD2C-inhibition is feasible in MPN patients, who carry a lower disease burden. If inhibition is well tolerated, JMJD2C represents a promising drug target in MPN possibly for synergistic use with JAK inhibitors, as Ernst et al. very recently demonstrated that JAK2V617F-mutated cells functionally depend on JMJD2C even during exposure to Ruxolitinib [13].
Data availability
Original datasets are available from the corresponding author on reasonable request.
References
Goerttler PS, Kreutz C, Donauer J, Faller D, Maiwald T, März E, et al. Gene expression profiling in polycythaemia vera: overexpression of transcription factor NF-E2. Br J Haematol. 2005;129:138–50.
Kaufmann KB, Gründer A, Hadlich T, Wehrle J, Gothwal M, Bogeska R, et al. A novel murine model of myeloproliferative disorders generated by overexpression of the transcription factor NF-E2. J Exp Med. 2012;209:35–50.
Jutzi JS, Bogeska R, Nikoloski G, Schmid CA, Seeger TS, Stegelmann F, et al. MPN patients harbor recurrent truncating mutations in transcription factor NF-E2. J Exp Med. 2013;210:1003–19.
Jutzi JS, Basu T, Pellmann M, Kaiser S, Steinemann D, Sanders MA, et al. Altered NFE2 activity predisposes to leukemic transformation and myelosarcoma with AML-specific aberrations. Blood. 2019;133:1766–77.
Marcault C, Zhao LP, Maslah N, Verger E, Daltro de Oliveira R, Soret-Dulphy J, et al. Impact of NFE2 mutations on AML transformation and overall survival in patients with myeloproliferative neoplasms. Blood. 2021;138:2142–8.
Wehrle J, Seeger TS, Schwemmers S, Pfeifer D, Bulashevska A, Pahl HL. Transcription factor nuclear factor erythroid-2 mediates expression of the cytokine interleukin 8, a known predictor of inferior outcome in patients with myeloproliferative neoplasms. Haematologica. 2013;98:1073–80.
Consortium TEP. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLOS Biol. 2011;9:e1001046.
Peeken JC, Jutzi JS, Wehrle J, Koellerer C, Staehle HF, Becker H, et al. Epigenetic regulation of NFE2 overexpression in myeloproliferative neoplasms. Blood. 2018;131:2065–73.
Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol J Am Soc Clin Oncol. 2011;29:1356–63.
Staehle HF, Pahl HL, Jutzi JS. The cross marks the spot: the emerging role of JmjC domain-containing proteins in myeloid malignancies. Biomolecules. 2021;11:1911.
Staehle HF, Heinemann J, Gruender A, Omlor AM, Pahl HL, Jutzi JS. Jmjd1c is dispensable for healthy adult hematopoiesis and Jak2V617F-driven myeloproliferative disease initiation in mice. PLOS One. 2020;15:e0228362.
Cheung N, Fung TK, Zeisig BB, Holmes K, Rane JK, Mowen KA, et al. Targeting aberrant epigenetic networks mediated by PRMT1 and KDM4C in acute myeloid leukemia. Cancer Cell. 2016;29:32–48.
Ernst P, Schnöder TM, Huber N, Perner F, Jayavelu AK, Eifert T, et al. Histone demethylase KDM4C is a functional dependency in JAK2-mutated neoplasms. Leukemia. 2022;36:1843–9.
Agger K, Nishimura K, Miyagi S, Messling JE, Rasmussen KD, Helin K. The KDM4/JMJD2 histone demethylases are required for hematopoietic stem cell maintenance. Blood. 2019;134:1154–8.
Wu Q, Young B, Wang Y, Davidoff AM, Rankovic Z, Yang J. Recent advances with KDM4 inhibitors and potential applications. J Med Chem. 2022;65:9564–79.
Acknowledgements
The authors sincerely thank Martina de Groot, Beate Krauss and Franziska Zipfel for expert technical assistance. HLP was funded by Deutsche Forschungsgemeinschaft (Pa611/5–3 and SFB992/TPB02). HSF was funded by Else Kröner-Fresenius-Stiftung (2021_EKEA.122). HFS is a fellow in the EXCEL clinician scientist program at the University of Freiburg Medical Faculty, funded by Else Kröner-Fresenius-Stiftung, granted to HLP (2016_Kolleg_15). NK and SBK are fellows in the IMMPACT clinician scientist program at the University of Freiburg Medical Faculty, funded by Deutsche Forschungsgemeinschaft (413517907). AMS, JCP, MEH, CK and HFS were funded by the MOTI-VATE scholarship program supported by Else Kröner-Fresenius-Stiftung. AMS was supported by a “Peter-Scriba Scholarship” of the Deutsche Gesellschaft für Innere Medizin. JSJ acknowledges funding from the German Research Foundation (DFG, JU 3104/2–1). JSJ is a Special Fellow of The Leukemia & Lymphoma Society (3415–22).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
AMS, JCP, GV, MEH, SBK, NK, CK, and HFS performed experiments. AMS, JCP, AG, JSJ, HLP and HFS designed experiments. AMS, JCP, GV, MEH, AG, CK, JSJ, HLP, HFS analyzed data. AMS, HLP and HFS wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Staehle, A.M., Peeken, J.C., Vladimirov, G. et al. The histone demethylase JMJD2C constitutes a novel NFE2 target gene that is required for the survival of JAK2V617F mutated cells. Leukemia 37, 919–923 (2023). https://doi.org/10.1038/s41375-023-01826-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01826-y