Abstract
T-cell dysregulation in chronic lymphocytic leukemia (CLL) associates with low response rates to autologous T cell-based therapies. How CLL affects antigen-specific T-cell responses remains largely unknown. We investigated (epi)genetic and functional consequences of antigen-specific T-cell responses in presence of CLL in vitro and in an adoptive-transfer murine model. Already at steady-state, antigen-experienced patient-derived T cells were skewed towards short-lived effector cells (SLEC) at the expense of memory-precursor effector cells (MPEC). Stimulation of these T cells in vitro showed rapid induction of effector genes and suppression of key memory transcription factors only in presence of CLL cells, indicating epigenetic regulation. This was investigated in vivo by following antigen-specific responses of naïve OT-I CD8+ cells to mCMV-OVA in presence/absence of TCL1 B-cell leukemia. Presence of leukemia resulted in increased SLEC formation, with disturbed inflammatory cytokine production. Chromatin and transcriptome profiling revealed strong epigenetic modifications, leading to activation of an effector and silencing of a memory profile through presence of CLL cells. Secondary challenge in vivo confirmed dysfunctional memory responses by antigen-experienced OT-I cells generated in presence of CLL. Altogether, we show that presence of CLL induces a short-lived effector phenotype and impaired memory responses by epigenetic reprogramming during primary responses.
Similar content being viewed by others
Introduction
The development of targeted small molecules such as BTK- and BCL-2 inhibitors has revolutionized the treatment of chronic lymphocytic leukemia (CLL). However, such treatments are not curative, resulting in a growing number of patients with double-class-resistant disease that lack effective salvage regimens [1]. Hence, in CLL an unmet need exists for additional therapeutic strategies with curative potential. Although CLL-directed T-cell therapy might be promising, there is a large gap between the efficacy observed in CLL and that observed in relapsed or refractory acute lymphoblastic leukemia, in which anti-CD19 CAR T cells induce complete remission in over 90% of cases [2, 3]. In CAR T-cell therapy of B-cell malignancies in general, and CLL specifically, expansion and persistence of adoptively transferred T-cell populations are considered a critical requirement for long-term tumor immunity without relapse [4]. Less differentiated (central) memory T cells are the main source of long-term persistence and seem crucial for successful adoptive cell therapy, in contrast to late-stage effector T cells that rapidly disappear following transfer [5].
Skewing of T-cell differentiation has been well described in CLL and is hallmarked by increasing absolute numbers of CD8+ T cells with progressive reductions of naive and accumulation of highly differentiated effector-memory subtypes [6]. The development of naive T cells into memory and effector cell populations upon an antigenic stimulus follows progressive epigenetic changes that instruct unique gene expression profiles and functions [7]. Ex vivo studies indicate that acquired T-cell dysfunction in CLL occurs through direct and indirect interaction with CLL cells [8,9,10]. However, whether CLL cells directly affect epigenetic programming and whether the impact of CLL on T-cell skewing depends on CLL-specific antigen recognition is currently unknown. It has been challenging to study the effect of CLL on a primary antigen-specific response due to difficulty to perform dynamic (long-term) T-cell studies on patient-derived samples in vitro. Studying antigen-specific T-cell responses in CLL can provide relevant clues how CLL influences T-cell functioning and skewing of non-CLL directed T cells, which may be of great importance to fully understand T-cell immunity in CLL patients.
Antigen-experienced CD8+ T cells from CLL patients were found to be skewed towards short-lived effector cells (SLECs) with reduction of memory features. Stimulation of these T cells resulted in the induction of an effector gene signature but only in presence of autologous CLL cells. To gain insights into the bystander effect of CLL on an antigen-specific immune response, we developed a combined murine model utilizing the CD45.2+ Eµ-T-cell leukemia-1 (TCL1) mouse model, that spontaneously develops an aggressive CD5+ CLL-like disease [11], with a CD45.1+ OT-I mouse model, which harbors CD8+ T cells that specifically recognize the ovalbumin (OVA) peptide SIINFEKL [12]. These data show that CLL-mediated changes in skewing and (recall) function are heavily epigenetically and transcriptionally regulated, which may provide clues for improvement of future T-cell-based therapies in CLL.
Materials and methods
Patients and controls
Peripheral blood mononuclear cells (PBMCs) were isolated and cryopreserved from whole blood of CLL patients or buffy coats of age-matched healthy donors (HD) (Table S1) as described before [8]. Written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki and the study was approved by the medical ethics committee at Academic Medical Center, Amsterdam, the Netherlands (ethics approval number 2013/159).
Mice
C57BL/6J mice were obtained from Charles River Laboratories and were 6–12 weeks old at the start of the experiment, all animals were sex-matched. OT-I transgenic (tg) and TCL1-tg mice were bred at the animal facility of the Amsterdam UMC and University of Rijeka and were bled monthly to monitor CLL development from the age of 8 months on. Upon full CLL development, mice were sacrificed and splenocytes were frozen for adoptive transfer (AT). Mice were housed under specific pathogen-free conditions in individually ventilated cages. All animal experiments were approved by the institutional animal experiments committees and executed according to the institutional, national, and European guidelines. More details regarding the ARRIVE guidelines can be found in the supplemental methods.
mCMV-OVA and determining viral titers
Murine cytomegalovirus (mCMV)-IE2-OVA or mCMV-N4 (both mCMV-OVA expressing the SIINFEKL peptide) and mCMV-Q4 (expressing SIIQFEKL) were generated as described previously [13,14,15]. For mCMV-N4 and mCMV-Q4, the bacterial artificial chromosome (BAC)-derived mCMV strain MW97.01 (in-house produced) has previously been shown to be biologically equivalent to the mCMV Smith strain (VR-1399; ATCC) For virus quantification, titers were determined on mouse embryonic fibroblasts by standard plaque assay.
In vivo experiments
C57BL/6J CD45.2 mice were intraperitoneally (i.p.) or intravenously (i.v.) injected with PBS (WT mice) or 20 × 106 TCL1 splenocytes (CLL mice). Mice were bled weekly to monitor CLL development. When mice fully developed CLL-like disease (>70% CD5+CD19+ of lymphocytes in blood), 50.000 OT-I cells were injected i.v., followed by i.p. injection of 0.1–0.2 × 106 PFU mCMV-OVA per mouse and sacrificed 7 days post-infection. Five independent experiments were performed using three different clones of TCL-1 cells.
For recall experiments, primary infection was done i.v. using mCMV-Q4. CD45.1+ OT-I cells 7 days after primary infection were isolated using CD8+ T Cell Isolation Kit (Miltenyi) and 50.000 OT-I cells were transferred into CD45.1+CD45.2+ WT mice. Five weeks after transfer mice were infected i.p. with mCMV-Q4 and sacrificed 6 days after secondary infection.
In vitro stimulation
Both human and mouse cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, supplemented with 10% fetal calf serum and 1% penicillin/streptomycin.
Human CLL T cells were stimulated using anti-CD3 (clone1XE) and anti-CD28 (clone 15E8) soluble antibodies for 2 days either in the original PBMC pool or after T-cell enrichment using the EasySep human T cell enrichment kit (StemCell technologies).
Mouse splenocytes were stimulated for 4 h with OVA-peptide (100 pg/mL or 10 ng/mL, SIINFEKL, Invivogen) or PMA (10 ng/mL, Sigma Aldrich) and ionomycin (1 µg/mL, Sigma Aldrich) in presence of Brefeldin A (10 µg/mL, Invitrogen), GolgiStop (BD Biosciences) and anti-CD107a. Cytokine production and degranulation were assessed via flow cytometry as described below.
Flow cytometry
Cells were stained with antibodies (details in supplemental materials and methods), acquired on BD FACS Canto or LSR Fortessa flow cytometer and analyzed with FlowJo v10.
Nanostring assay
Human CD8+ T cells were FACS-sorted from the culture with CLL cells and the enriched T-cell condition using the SH800 Cell Sorter (Sony), the cells were pelleted and total RNA was isolated using the RNeasy mini kit (Qiagen) following the manufacturers protocol. RNA was prepared with the nCounter Low RNA input amplification kit (NanoString), samples were analyzed on the nCounter SPRINT profiler using the CAR-T panel (NanoString). For further details see supplemental materials and methods.
ATAC sequencing and gene expression profiling
Assay for transposase accessible chromatin sequencing (ATACseq) libraries of OT-I cells were generated following the Omni-ATAC protocol [16]. For RNA sequencing (RNSseq) total RNA from FACS-sorted OT-I cells was isolated using the RNeasy micro kit (Qiagen) following the manufacturers protocol. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was used for gene expression analysis of a selection of genes. For further details see supplemental materials and methods.
Statistical analysis
Data was checked for normality by a Shapiro-Wilk test. P values were calculated using two-sided unpaired t tests or ordinary one-way ANOVA (followed by Bonferroni’s post hoc test). Statistical analysis was performed using Graphpad PRISM version 9.1.0 (significance P < 0.05). Data are presented as mean ± standard deviation (SD) unless indicated differently.
Results
CLL cells induce in vitro effector skewing accompanied by an effector transcriptional signature
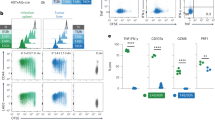
Compared to age-matched HD, T cells of CLL patients were skewed towards an effector/memory phenotype demonstrated by a reduced proportion of naïve cells and increased effector cells as previously shown [9, 17] (Fig. 1A, gating strategy in Fig. S1). Non-naive CD8+ cells were gated based on KLRG1 and CD127 expression in order to differentiate between memory precursor effector cells (MPECs), cells that differentiate into long-lived memory T cells, and SLECs, cells that usually contract after infection, and only marginally contributing to memory formation [18]. T cells from CLL patients were skewed towards SLECs at the expense of MPECs (Fig. 1B). Expression of CD45RA, a marker of differentiated TEMRA cells, was increased while CD28 was decreased on SLECs from CLL patients as compared to HD. Total CD8+ T cells of CLL patients showed reduced expression of naïve/memory markers CD27 and CD28 (Fig. 1C).
A Differentiation of T cells from CLL patients and age-matched healthy donors (HD). Cells were gated as naïve CD8+CD45RA+CD27+; memory CD8+CD45RA−CD27+; effector CD8+CD27− directly after thawing PBMCs (N = 12 per group). B From the non-naïve CD8+ cells, memory precursor effector cells (MPEC; KLRG1-CD127+) and short-lived effector cells (SLEC; KLRG1+CD127−) were gated for CLL patients and HD. C Within total CD8+, MPEC, and SLEC T cells the expression of CD45RA, CD27, and CD28 is depicted. D CD25 expression on CD8+ T cells after 2 days of culture, either unstimulated (−) or stimulated with soluble αCD3/αCD28 antibodies. Conditions are HD PBMCs or CLL T cells in the presence or absence of autologous CLL cells (N = 3–4). E Volcano plot depicting the differentially expressed genes in black (FDR < 0.1) and highlighting relevant T-cell genes. The P value was calculated by unpaired T-test (A, B) one-way ANOVA (followed by Bonferroni’s post hoc test) (C) or paired t test (D). Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
So far, these data were obtained from samples directly after thawing. Next, we studied responses upon T-cell receptor stimulation presence or absence of CLL cells. To prevent bias by subset skewing between healthy donors and CLL patients at baseline, we applied a paired experimental design using only CLL T cells stimulated in the presence or absence of their autologous CLL cells, taking healthy donor PBMCs along for reference. After 2 days of T-cell stimulation, presence of CLL cells strongly reduced T-cell activation in comparison to age-matched healthy donors measured by CD25 expression (Fig. 1D) as earlier described [8]. Transcriptional analysis of the CD8+ T cells from both culture conditions demonstrated clear separation in a principal component analysis (Fig. S2A). Presence of CLL cells induced an effector gene signature in the T cells (Fig. 1E) and led to reduced expression of memory-associated transcription factor BATF3 [19]. A Gene Set Enrichment Analysis (GSEA) using eight pathways curated by Hofland et al. demonstrated significant enrichment of the effector gene set in the culture with CLL cells [20] (Fig. S2B; Table S2).
Although T-cell dysfunction in conjunction with transcriptional and phenotypical skewing towards (short-lived) effector cells could be observed in this system, the full impact of CLL cells on antigen-specific T-cell differentiation and function cannot be studied effectively in vitro. We, therefore, implemented mouse models to allow more detailed characterization of antigen-specific T-cell responses in the presence of CLL.
TCL1 leukemia skew antigen-specific T cells towards a short-lived effector phenotype during an acute infection
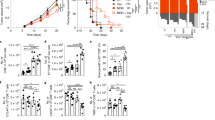
Besides skewing towards an effector/memory phenotype, T cells of CLL patients have an inverted CD4/CD8 ratio, compared to healthy donors [9, 17, 21]. Concordantly, upon the development of CLL-like disease in mice following adoptive transfer (AT) of leukemic TCL1 splenocytes, this skewing and CD4/CD8 reversal was also present and a trend towards increased PD-1 expression on CD8+ T cells from CLL mice could be observed [22, 23] (Fig. S3A–C). In order to study an acute primary T-cell response in the presence of CLL-like disease, mice were weekly screened following AT. Upon development of leukemia and associated native T-cell skewing (Fig. S3A), OVA-specific OT-I cells with a different congenic marker (CD45.1) were injected followed by infection with mCMV-OVA (Fig. 2A). Age-matched mice without AT were used as control. At the peak of infection (day seven) animals were sacrificed and OT-I T-cell differentiation was assessed in various organs (gating strategy in Fig. S4). As the spleen in this model mostly resembles the lymph node environment in patients [11], we focus on spleen unless stated otherwise. Both absolute numbers, as well as percentage of splenic OT-I CD8+ cells, were increased in TCL1 mice (Fig. 2B) and virtually all OT-I cells differentiated into effector cells (Fig. 2C). In line with this, viral clearance was not different when TCL1 or WT mice received OT-I cells (Fig. 2D). However, TCL1-derived OT-I cells showed increased SLEC skewing and decreased MPECs in both spleen and blood (Fig. 2E and F, S5A). No MPEC/SLEC skewing was seen in mesenteric and peripheral LNs, in line with no to low presence of leukemic cells in in this model [11] (Fig. S5B). Together, these data show that CLL-like disease skews (antigen-specific) T-cell responses towards a SLEC phenotype at compartments with high presence of leukemia cells, indicating impaired early memory formation.
A Schematic overview of experimental setup. B Magnitude of response measured in spleen on day 7 as percentage (left panel) or absolute number (right panel) of OT-I cells (CD8+CD45.1+). (n = 5 per group) C Differentiation of OT-I cells into effector cells in spleen on day 7. (n = 5 per group). D Determination of viral titers derived from liver samples in CLL and WT mice (n = 5 per group) E, F Differentiation of OT-I cells into MPECs (KLRG1-CD127+) and SLECs (KLRG1+CD127−) in spleen on day 7. (n = 5 per group). The P value was calculated by one-way ANOVA (followed by Bonferroni’s post hoc test) A, B or unpaired t test (D, E, F, G). Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001 ****P < 0.0001. CLL mice are mice that received TCL1 splenocytes through AT and data are representative of five independent experiments.
Restimulation of TCL1-derived OT-I cells results in dysfunctional cytokine production and degranulation
To determine whether antigen-experienced OT-I cells derived from TCL1 mice show signs of a dysfunctional recall response following infection, splenocytes were restimulated specifically with the OVA-peptide or non-specifically using PMA/Ionomycin in vitro. Upon specific restimulation, OT-I cells derived from TCL1 mice had lower capacity to produce IFNγ, TNFα, IL-2 and had lower degranulation capacity (CD107a) (Fig. 3A, S6A). Also, a trend towards lower Granzyme B could be observed (Fig. S6A). Non-specific stimulation also resulted in lower amounts of IFNγ, TNFα, CD107a, and IL-2 in the CLL-derived OT-I cells (Fig. 3B, Fig. S6B). We next looked for differences in cytokine expression specifically in the SLEC population (KLRG1BRIGHT). Both IFNγ and CD107a expression were lower in OT-I SLECS from TCL1 mice (Fig. 3C). In contrast, the endogenous CD8+ compartment of CLL mice. which have been in contact with TCL1 cells during the entire experiment, showed increased IFNγ production, equal TNFα levels and reduced CD107a expression upon stimulation with PMA/ionomycin (Fig. S6C), compared to WT mice. This is similar to what is described for CLL patients [8, 9]. In conclusion, cytokine production and degranulation in OT-I cells from TCL1 mice are impaired upon restimulation even within KLRG1 + cells.
Expression of IFNγ (left panels), TNFα (middle panels), or CD107a (right panels) by total OT-I cells after restimulation with A 100 pg/mL SIINFEKL peptide or B PMA/ionomycin. C Expression of IFNγ (left panels), TNFα (middle panels) or CD107a (right panels) by KLRG1 + OT-I cells after restimulation with 100 pg/mL SIINFEKL peptide (n = 5 per group). The P values were calculated using unpaired t test (A–C). Data are presented as mean ± SD. *P < 0.05; ***P < 0.001. CLL mice are mice that received TCL1 splenocytes through adoptive transfer and data are representative of five independent experiments.
TCL1-derived OT-I cells express lower levels of memory-related transcription factors
Transcription factors play a major role in fate decision between MPECs and SLECs [24]. We assessed expression of T-box expressed in T cells (T-BET), Eomesodermin (EOMES), and B-cell lymphoma 6 (BCL-6). T-BET is associated with SLEC formation, while EOMES and BCL-6 drive memory development and MPEC differentiation [15, 25, 26]. In concordance with the SLEC phenotype, TCL1-derived OT-I cells showed increased T-BET expression, and decreased EOMES and a trend towards lower BCL-6 expression (Fig. 4A). Development of T-cell exhaustion has been described in CLL patients and in CLL mouse models [9, 22, 27]. We, therefore, studied whether the observed skewing and dysfunction were a derivative of exhaustion. In vivo activated OT-I cells did express the exhaustion-associated markers PD-1, TIM-3, CD101, and CD200R [28], but expression of these markers was lower in the OT-I cells obtained from the TCL1 mice compared to WT mice (Fig. 4B). No differences were found in the expression of CTLA4 (Fig. S7A).
A Expression of T-BET (left panel), EOMES (middle panel), and BCL-6 (right panel) on OT-I cells derived from spleen on day 7 (n = 5 per group). B Expression of PD-1, TIM-3, CD101 and CD200R on OT-I cells derived from spleen on day 7 (n = 5 per group). The P values were calculated using unpaired t test (A–C). Data are presented as mean ± SD. *P < 0.05; ***P < 0.001; ****P < 0.0001. CLL mice are mice that received TCL1 splenocytes through adoptive transfer and data are representative of five independent experiments.
In conclusion, increased expression of T-BET and decreased expression of EOMES and BCL-6 are likely important regulators of the observed SLEC skewing, while no signs of increased exhaustion could be find at the protein level.
Transcriptional program of OT-I cells reflects effector skewing and reduced expression of memory genes in CLL mice
We next performed RNAseq to study transcriptional differences between the antigen-experienced OT-I cells from WT and TCL1 mice. Differential expression analysis revealed 182 genes significantly differentially expressed (FDR < 0.05 and fold change > 2) whereby 123 genes were higher expressed in OT-I from TCL1 mice (Fig. 5A). Expression of Klrg1 was higher in TCL1 and Il7r (CD127) expression was reduced in TCL1 OT-I cells, in line with FACS data. Moreover, expression of numerous known effector genes such as Ccl4, Id2, and Ifng was increased in TCL1-derived OT-I cells (Fig. 5B, left panel) whilst key memory genes such as Id3, Ccr7 and Sell (encoding CD62L) were clearly downregulated in the TCL1 condition (Fig. 5B, right panel). Gene set enrichment analysis showed that OT-I cells from TCL1 mice resemble the transcriptional program of effector rather than memory CD8+ cells (Fig. 5C, top panel). Additionally, WT OT-I cells were enriched for genes upregulated in IL7R high effector CD8+ cells (Fig. 5C, bottom panel). Expression of key effector and memory genes that were found to be differentially expressed by RNAseq analysis was confirmed by RT-qPCR (Fig. S8).
A Volcano plot with each dot representing a gene, whereby black dots are regions with FDR < 0.05 and fold change > 2 B Log2 fold change of relevant effector and memory genes when comparing OT-I from CLL compared to WT. C Gene set enrichment analysis on normalized counts (DESeq2 median of ratios normalization) using MSigDB Immunological Signature gene sets (c7.v7.4). Genes are ranked by differential expression in CLL vs WT on the x-axis: red, up in CLL; blue, up in WT. Curves (green) indicate cumulative enrichment quantified by enrichment score on the y-axis. Tick marks on the x-axis correspond to the rank of genes in the gene set. Top panel is gene set M3041 [48] and bottom panel gene set M5771 [44]. D Gene set enrichment analysis with public data by Hanna et al [29]. The Y-axis depicts the comparison and whether it contains the genes of which expression increased (UP) or decreased (DOWN) in the given comparison. PD1HI = CD8 PD1 high expressing T cells; PD1INT = CD8 PD1 intermediate expressing T cells; MEM = memory CD8 T cells; NAÏVE = naïve CD8 T cells; NES = normalized enrichment score. CLL mice are mice that received TCL1 splenocytes through adoptive transfer.
When comparing our data to publicly available data on CD8+ T cells in the TCL1 mouse model, we found that OT-I cells from TCL1 mice were more similar to PD1-high and PD1-intermediate cells as described by Hanna et al [29]. (Fig. 5D). Even though at protein level we did not find clear indications of exhaustion, these data demonstrated that CLL-like disease induced an (early) exhaustive signature in OT-I cells at the transcriptional level.
TCL1-derived OT-I cells have altered chromatin accessibility and show reduced accessibility of binding motifs for memory-related transcription factors
To unravel underlying gene regulation leading to the transcriptional differences and to assess whether CD8+ T-cell differentiation, including SLEC and MPEC formation is epigenetically regulated in this model [30], ATACseq analysis was performed on splenic OT-I cells seven days after infection in order to measure chromatin accessibility. A clear separation between the WT and TCL1 conditions was observed in a principal component plot (Fig. S9A). In total 2615 genomic regions were significantly differential accessible (FDR < 0.05) with 1620 regions more accessible in TCL1 OT-I cells (Fig. 6A, B). In correspondence with SLEC skewing (e.g., KLRG1+) and higher gene expression of Klrg1 in the OT-I from TCL1 mice, Klrg1 displayed more accessible chromatin in the TCL1 condition (Fig. 6C, D). In line with the hypothesis of impaired memory formation in CLL, chromatin of two enhancer regions of Ccr7, a known memory marker on T cells [18], was less accessible in TCL1 mice (Fig. 6C, D). When comparing these data to chromatin accessibility in CD8+ subsets as described by Scott-Browne et al. [30], we found depletion of naive- and enrichment of effector-specific accessible regions in the OT-I cells from TCL1 mice (Fig. S9B).
A Heatmap depicting the normalized counts of the differentially accessible regions using hierarchical clustering on columns (Euclidian distance method). B Volcano plot with each dot representing a region, whereby black dots are regions with FDR < 0.05 and fold change > 2. C UCSC genome browser graphs of chromatin profiles near Klrg1 and Ccr7. D Fold change and FDR of regions 1 and 2 of Klrg1 and Ccr7. E Transcription motif enrichment analysis on peaks significantly more accessible in CLL and WT. F Heatmap showing normalized ATACseq counts of selected promoter regions containing an ETS1 motif. CLL mice are mice that received TCL1 splenocytes through adoptive transfer.
To infer transcription factors associated with differences in chromatin landscape, HOMER’s motif analysis was performed on regions significantly more accessible in TCL1 and in WT (Fig. 6E). Clear enrichment of ETS-family transcription factor motifs was observed in OT-I from TCL1 mice, with ETS1 as most significant hit. More accessible chromatin regions in TCL1 OT-I close to promoters (<3 kb) that contain an ETS1 motif were annotated to effector-related genes such as Klrg1, Lamp1 (encoding CD107a), and Ifng (Fig. 6F, Table S5) and transcription factors Fosb, Id2 and Fos, known for establishing an effector phenotype in CD8+ T cells [31,32,33].
In contrast, more accessible chromatin of antigen-experienced OT-I cells from WT mice showed enrichment for high-mobility group transcription factor motifs, more specifically the highly homologous TCF/LEF family members (Fig. S9C, Table S6). Tcf7 (encoding TCF1) is involved in self-renewal of T cells and its role in CD8+ T-cell memory has been described extensively [34, 35], also in cooperation with LEF1 [36, 37]. Taken together, the chromatin accessibility profiles and corresponding transcription factor motifs in OT-I from TCL1 mice reflect an effector skewing at the expense of memory features.
Chromatin accessibility results overlap with gene expression data on several crucial parts (Fig. S10A, Tables S7, 8). Genes that showed more accessible chromatin in TCL1 OT-I such as Klrg1, Fos, Ifng, Ccl4, Id2, and Entpd1 (encoding CD39) were also higher expressed in TCL1 OT-I and contained open ETS1 motifs in their promoters (Fig. S10B). This gene profile largely overlaps with immune response genes that specifically recruit the Ets1 transcription factor upon activation in CD8+ T cells as described [38], indicating a central role for ETS1 in establishing the effector transcriptional program. In WT OT-I Ccr7 and Tcf7 were more accessible and higher expressed, which, together with enrichment for TCF-family motifs, indicates a TCF-centered gene regulatory network.
Together these data confirm an epigenetic and transcriptional program skewed towards effector phenotype in TCL1-derived OT-I cells, implying epigenetically regulated dysregulated memory formation.
TCL1-derived memory T cells are defective upon secondary infection
To determine if in vivo memory function was affected by CLL, as hypothesized based on flow cytometry, epigenetic and transcriptional profiles, recall responses were determined as depicted in Fig. 7A. Six days after the secondary infection with mCMV-OVA, decreased percentage of OT-I cells of total CD8+ cells was observed in mice that received TCL1-derived OT-I cells, coinciding with decreased absolute OT-I counts (Fig. 7B). Most OT-I cells differentiated into effector cells (CD62L−CD44+) and no difference in the percentage of memory cells (CD62L+CD44-) was observed (Fig. S11A). Despite lower persistence of the TCL1-derived OT-I cells, they showed increased production of effector cytokines IFNγ, TNFα, and Granzyme B, while no difference in CD107a was observed (Fig. 7C, Fig. S11B) and no differences in T-BET or EOMES could be observed (Fig. S11C). Together these data indicate decreased persistence and proliferation of antigen-experienced OT-I cells following a secondary infection, implying impaired memory responses.
A Schematic overview of experimental setup. B Magnitude of response measured in spleen on day 7 after infection as (B) percentage of OT-I cells of total CD8+ cells or absolute number of OT-I cells (CD8+CD45.1+CD45.2−). C Expression of IFNγ, TNFα, Granzyme B by OT-I cells from spleen on day 7 after secondary infection followed by restimulation with 10 ng/mL SIINFEKL peptide. (n = 4–5 per group) The P value was calculated by unpaired t test (B–I). Data are presented as mean ± SD. *P < 0.05. CLL mice are mice that received TCL1 splenocytes through adoptive transfer and data are representative of one independent experiments.
Discussion
Our study demonstrates that the presence of CLL cells results in skewing towards a dysfunctional short-lived effector phenotype following a non-CLL antigen-specific T-cell response. Skewing is accompanied by an altered epigenomic and transcriptomic regulation of key effector and memory genes, leading to impaired CD8+ T-cell memory formation. In vitro modeling of response to a single antigen cannot fully recapitulate natural human infections, but these results imply that CLL-like disease hampers optimal memory T-cell formation against specific antigens. This impacts responses to viral infections in patients and might have implications for the efficacy of cellular therapy. Although mechanistic studies on T-cell memory against COVID-19 are lacking, recent reports showing reduced cellular immune responses to the BNT162b2 mRNA COVID-19 vaccine in most CLL patients do indicate diminished anti-viral responses in this disease [39, 40].
A key question is how CLL or TCL1 cells impair T-cell function mechanistically. Although effector skewing was observed, these cells had impaired ability to perform effector functions such as IFNγ production, 7 days after primary infection. Reduced cytokine expression could be a marker of exhaustion [41]. Despite our observations that immune checkpoint expression was not upregulated in OT-I cells of TCL1-mice at the protein level, we did find enrichment of exhaustion-related genes when comparing our data to publicly available data on exhausted CD8+ cells from the same TCL1 model. These observations are indicative of an early exhaustive state without the classical markers being upregulated (yet) on the cell surface. Another scenario is that CLL-like disease disrupts the normal cellular architecture within secondary lymphoid organs [42]. CLL is well known to accumulate in lymph nodes and spleen and may impact antigen presentation and the chemokine and cytokine environment. Alternatively, CLL cells may be directly responsible for defective T-cell activation. These cells have impaired antigen-presenting capacity which correlates with low expression levels of costimulatory markers such as CD80 and CD86 [43].
Epigenomic analyses are widely employed to understand T-cell dysfunction, yet how CLL influences the T-cell epigenome is still unknown [7]. We show extensive differences in OT-I cells from WT and TCL1 mice regarding chromatin accessibility, establishing that the presence of leukemic cells during infection alters the T-cell epigenome. Differential chromatin accessibility profiles, transcription factor motif enrichment, and altered gene expression are all indicative of skewing towards effector differentiation in TCL1 mice. Whilst the role of T-BET is well established in SLEC formation [44], we demonstrate ETS-family transcription factors might also be involved in the CD8+ effector skewing. Recently, Zhong et al. described that ETS1 acts as a “housekeeper” in CD8+ T cells and its function is highly dependent on recruitment of a variety of transcription factors and other co-factors. Furthermore, in a subset of immune response genes, highly overlapping with our effector gene signature, ETS1 is specifically recruited upon activation [38]. Others have described that exhausted CD8+ T cells show enrichment of ETS binding sites compared to stem-like memory cells [35]. How CLL affects the role of ETS1 in CD8+ T cells remains to be elucidated. In addition, the data imply a role for transcriptional regulators Id2 and Id3, as Id2 had more open chromatin and was higher expressed in TCL1-derived OT-I cells, whilst Id3 was higher expressed in WT OT-I. Interplay between these two factors connects with cell-fate determination as Id2 is essential for short-term survival of effector cells and its loss leads to enhanced memory formation coinciding with increased Eomes and Tcf7 expression [33, 45]. On the other hand, Id3 is enriched in MPECs and required for their long-term survival [46]. This is in agreement with our data where we observe reduced BATF3 expression in the patient samples, an essential transcription factor for the transition of CD8+ effector to memory cells [19], and depletion of motifs for the TCF/LEF family transcription factors, well established for their role in T-cell memory [32] in TCL1 mice. It was recently demonstrated that TCF1 and LEF1 highly impact persistence of CAR T cells in acute lymphoblastic leukemia and CLL [47]. The reduced expression of Tcf7 in CLL OT-I cells is therefore a likely cause of the observed impaired persistence.
Since memory properties are critical for autologous T-cell therapy, these data provide clues for future T-cell-based therapies for CLL. Our results show that exposure of T cells to a CLL-like environment during priming impairs memory recall during secondary antigen-specific response. Parallels between memory formation after acute infection or upon CAR T-cell treatment can be drawn as both depend on the capacity for self-renewal and T-cell persistence. Specifically, the role of TCF1 and LEF1 seems crucial in both models [4, 47]. In CLL patients it was determined that remission corresponded to elevated numbers of memory-like cells before CAR T-cell production as well as a memory-related transcriptional profile in the pre-infusion product [4]. Overall, our model is suitable for increasing understanding of key T-cell dynamics and guiding future research into this clinically relevant problem.
In conclusion, our results demonstrate that CLL induces skewing of antigen-specific T cells towards a dysfunctional short-lived phenotype during acute infection. This is accompanied by the remodeling of chromatin accessibility, resulting in impaired memory formation. Since T-cell dysfunction in the TCL1 model and CLL patients is highly similar, this implies CLL might have a similar effect during infection in patients.
Data availability
All data are available upon reasonable request. RNAseq and ATACseq raw and processed data have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE185387. Code is available upon reasonable request.
References
Lew TE, Lin VS, Cliff ER, Blombery P, Thompson ER, Handunnetti SM, et al. Outcomes of patients with CLL sequentially resistant to both BCL2 and BTK inhibition. Blood Adv. 2021;5:4054–8.
Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–17.
Siddiqi T, Soumerai JD, Dorritie KA, Stephens DM, Riedell PA, Arnason JE, et al. Phase 1 TRANSCEND CLL 004 study of lisocabtagene maraleucel in patients with relapsed/refractory CLL or SLL. Blood. 2022;139:1794–806.
Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018;24:563–71.
McLellan AD, Ali Hosseini Rad SM. Chimeric antigen receptor T cell persistence and memory cell formation. Immunol Cell Biol. 2019;97:664–74.
Man S, Henley P. Chronic lymphocytic leukaemia: the role of T cells in a B cell disease. Br J Haematol. 2019;186:220–33.
Peters FS, Strefford JC, Eldering E, Kater AP. T-cell dysfunction in chronic lymphocytic leukemia from an epigenetic perspective. Haematologica. 2021;106:1234–43.
van Bruggen JAC, Martens AWJ, Fraietta JA, Hofland T, Tonino SH, Eldering E, et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8(+) T cells and impede CAR T-cell efficacy. Blood. 2019;134:44–58.
Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121:1612–21.
Görgün G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Investig. 2005;115:1797–805.
Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99:6955–60.
Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T-cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27.
Dekhtiarenko I, Ratts RB, Blatnik R, Lee LN, Fischer S, Borkner L, et al. Peptide processing is critical for T-cell memory inflation and may be optimized to improve immune protection by CMV-based vaccine vectors. PLoS Pathog. 2016;12:e1006072.
Lemmermann NA, Gergely K, Bohm V, Deegen P, Daubner T, Reddehase MJ. Immune evasion proteins of murine cytomegalovirus preferentially affect cell surface display of recently generated peptide presentation complexes. J Virol. 2010;84:1221–36.
Kavazovic I, Han H, Balzaretti G, Slinger E, Lemmermann NAW, Ten Brinke A, et al. Eomes broadens the scope of CD8 T-cell memory by inhibiting apoptosis in cells of low affinity. PLoS Biol. 2020;18:e3000648.
Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–62.
Mackus WJ, Frakking FN, Grummels A, Gamadia LE, De Bree GJ, Hamann D, et al. Expansion of CMV-specific CD8+CD45RA+CD27- T cells in B-cell chronic lymphocytic leukemia. Blood. 2003;102:1057–63.
Martin MD, Badovinac VP. Defining memory CD8 T cell. Front Immunol. 2018;9:2692.
Ataide MA, Komander K, Knopper K, Peters AE, Wu H, Eickhoff S, et al. BATF3 programs CD8(+) T cell memory. Nat Immunol. 2020;21:1397–407.
Hofland T, de Weerdt I, Endstra S, Jongejan A, Platenkamp L, Remmerswaal EBM, et al. Functional differences between EBV- and CMV-specific CD8(+) T cells demonstrate heterogeneity of T cell dysfunction in CLL. Hemasphere. 2020;4:e337.
Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Investig. 2008;118:2427–37.
McClanahan F, Riches JC, Miller S, Day WP, Kotsiou E, Neuberg D, et al. Mechanisms of PD-L1/PD-1-mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Emicro-TCL1 CLL mouse model. Blood. 2015;126:212–21.
McClanahan F, Hanna B, Miller S, Clear AJ, Lichter P, Gribben JG, et al. PD-L1 checkpoint blockade prevents immune dysfunction and leukemia development in a mouse model of chronic lymphocytic leukemia. Blood. 2015;126:203–11.
Amsen D, Backer RA, Helbig C. Decisions on the road to memory. Adv Exp Med Biol. 2013;785:107–20.
Mathieu C, Beltra JC, Charpentier T, Bourbonnais S, Di Santo JP, Lamarre A, et al. IL-2 and IL-15 regulate CD8+ memory T-cell differentiation but are dispensable for protective recall responses. Eur J Immunol. 2015;45:3324–38.
Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177:7515–9.
Llao Cid L, Hanna BS, Iskar M, Roessner PM, Ozturk S, Lichter P, et al. CD8(+) T-cells of CLL-bearing mice acquire a transcriptional program of T-cell activation and exhaustion. Leuk Lymphoma. 2020;61:351–6.
Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1(+) stem-like CD8(+) T cells during chronic infection. Immunity. 2019;51:1043–58.e4.
Hanna BS, Llaó-Cid L, Iskar M, Roessner PM, Klett LC, Wong JKL, et al. Interleukin-10 receptor signaling promotes the maintenance of a PD-1(int) TCF-1(+) CD8(+) T cell population that sustains anti-tumor immunity. Immunity. 2021;54:2825–41.e10.
Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, et al. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity. 2016;45:1327–40.
Papavassiliou AG, Musti AM. The multifaceted output of c-Jun biological activity: focus at the junction of CD8 T cell activation and exhaustion. Cells. 2020;9:2470.
Zhou S, Cerny AM, Fitzgerald KA, Kurt-Jones EA, Finberg RW. Role of interferon regulatory factor 7 in T cell responses during acute lymphocytic choriomeningitis virus infection. J Virol. 2012;86:11254–65.
Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, et al. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol. 2013;190:4585–94.
Raghu D, Xue HH, Mielke LA. Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol. 2019;40:1149–62.
Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin JX, et al. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci USA. 2019;116:14113–8.
Zhou X, Xue HH. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol. 2012;189:2722–6.
Zhao X, Shan Q, Xue HH. TCF1 in T cell immunity: a broadened frontier. Nat Rev Immunol. 2022;22:147–57.
Zhong Y, Walker SK, Pritykin Y, Leslie CS, Rudensky AY, van der Veeken J. Hierarchical regulation of the resting and activated T cell epigenome by major transcription factor families. Nat Immunol. 2022;23:122–34.
Mellinghoff SC, Robrecht S, Mayer L, Weskamm LM, Dahlke C, Gruell H, et al. SARS-CoV-2 specific cellular response following COVID-19 vaccination in patients with chronic lymphocytic leukemia. Leukemia. 2022;36:562–5.
Itchaki G, Rokach L, Benjamini O, Bairey O, Sabag A, Vernitsky H, et al. Cellular immune responses to BNT162b2 mRNA COVID-19 vaccine in patients with chronic lymphocytic leukemia. Blood. 2021;138:638. Supplement 1
Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. Defining ‘T cell exhaustion’. Nat Rev Immunol. 2019;19:665–74.
Herishanu Y, Katz BZ, Lipsky A, Wiestner A. Biology of chronic lymphocytic leukemia in different microenvironments: clinical and therapeutic implications. Hematol Oncol Clin North Am. 2013;27:173–206.
Ranheim EA, Kipps TJ. Activated T cells induce expression of B7/BB1 on normal or leukemic B cells through a CD40-dependent signal. J Exp Med. 1993;177:925–35.
Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–95.
Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–9.
Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12:1230–7.
Chen GM, Chen C, Das RK, Gao P, Chen CH, Bandyopadhyay S, et al. Integrative bulk and single-cell profiling of premanufacture t-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov. 2021;11:2186–99.
Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103:3304–9.
Acknowledgements
The authors would like to thank the Core Facility Genomics at the Amsterdam UMC for assistance in troubleshooting and sequencing the ATAC libraries and Dr. Maria Themeli for valuable scientific discussions. This work was supported by the Netherlands Organization for Scientific Research (NWO)/Netherlands Organization for Health Research and Development (ZonMw) VIDI grant (#91715337) and European Research Council (ERC) Consolidator grant (BOOTCAMP; 864815).
Author information
Authors and Affiliations
Contributions
AWJM, EE, GJWvdW, FMW, FSP, and APK designed research. AWJM, IK, MK, GJWvdW, FMW, FSP, and SMP performed research. AWJM, IK, AJ, PDM, FMW, FSP analyzed data. AWJM and FSP designed the figures. RA provided samples. AWJM, FSP, and APK wrote the paper. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
GJWvdW is an employee of Genmab. All other authors declare that they have no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Martens, A.W.J., Kavazović, I., Krapić, M. et al. Chronic lymphocytic leukemia presence impairs antigen-specific CD8+ T-cell responses through epigenetic reprogramming towards short-lived effectors. Leukemia 37, 606–616 (2023). https://doi.org/10.1038/s41375-023-01817-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01817-z