Abstract
Acute myeloid leukemia (AML) is characterized by complex molecular alterations and driver mutations. Elderly patients show increased frequencies of IDH mutations with high chemoresistance and relapse rates despite recent therapeutic advances. Besides being associated with global promoter hypermethylation, IDH1 mutation facilitated changes in 3D DNA-conformation by CTCF-anchor methylation and upregulated oncogene expression in glioma, correlating with poor prognosis. Here, we investigated the role of IDH1 p.R132H mutation in altering 3D DNA-architecture and subsequent oncogene activation in AML. Using public RNA-Seq data, we identified upregulation of tyrosine kinase PDGFRA in IDH1-mutant patients, correlating with poor prognosis. DNA methylation analysis identified CpG hypermethylation within a CTCF-anchor upstream of PDGFRA in IDH1-mutant patients. Increased PDGFRA expression, PDGFRA-CTCF methylation and decreased CTCF binding were confirmed in AML CRISPR cells with heterozygous IDH1 p.R132H mutation and upon exogenous 2-HG treatment. IDH1-mutant cells showed higher sensitivity to tyrosine kinase inhibitor dasatinib, which was supported by reduced blast count in a patient with refractory IDH1-mutant AML after dasatinib treatment. Our data illustrate that IDH1 p.R132H mutation leads to CTCF hypermethylation, disrupting DNA-looping and insulation of PDGFRA, resulting in PDGFRA upregulation in IDH1-mutant AML. Treatment with dasatinib may offer a novel treatment strategy for IDH1-mutant AML.

Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) describes a myeloid disorder that is characterized by oncogenic transformation and uncontrolled clonal expansion of poorly differentiated cells of myeloid origin [1]. Among the adult population, AML is one of the most common types of leukemia [2]. On the molecular level, AML represents a largely heterogeneous disease, characterized by complex patterns of molecular alterations such as gene fusions or rearrangements together with somatically acquired driver mutations [3]. The advancing knowledge of molecular pathogenesis in AML led to the proposal of molecular classifications currently dividing AML into 11 subgroups by cytogenetic aberrations and different prognostic markers [4]. Frequently mutated genes belong to numerous functional categories including signaling genes (FLT3, KRAS, NRAS), nucleophosmin (NPM1), myeloid transcription factors (RUNX1, CEBPA), chromatin regulators (ASXL1) or DNA methylation-associated genes and epigenetic regulators (DNMT3A, TET2, IDH1/2) [3].
In younger adults, IDH1 mutations occur in 6-10% of AML cases, whereas elderly AML patients show a significantly higher IDH1 mutation frequency of ~17% accompanied by global DNA hypermethylation [5]. IDH1 and IDH2 catalyze the conversion of isocitrate to α-ketoglutarate (α-KG) in the cytoplasm (IDH1) or the mitochondria (IDH2) as part of the citrate cycle. α-KG further activates enzymes of the ten-eleven translocation (TET) family in the nucleus that are essential mediators of DNA demethylation and subsequent transcriptional activation of target genes [1, 6,7,8]. The most common mutation reported for IDH1 is p.R132H, resulting from a G to A base shift that leads to an amino acid shift from arginine at position 132 to histidine and a subsequent conformation change in the IDH1 protein [6]. As a result, an alternative oncometabolite 2-hydroxygluterate (2-HG) is generated, which competes with α-KG for binding at the TET enzymes while it lacks the ability to activate them. Subsequently, TET enzyme activity is inhibited, resulting in inhibition of demethylation and therefore a hypermethylation phenotype [1, 5, 9]. Studies have shown that such hypermethylation is often found at promoters of genes involved in cell differentiation, connecting the occurrence of IDH mutations to the inhibition of myeloid differentiation [7]. The recent approval of the therapeutic drugs ivosidenib (IDH1) and enasidenib (IDH2) by the Food and Drug Administration (FDA) has been an important advance in the treatment of patients with AML patients containing IDH mutations [10]. However, resistance and relapse still remain a problem to be addressed [11].
In addition to a differentiation block mediated by hypermethylated gene promotors, recent research suggests an alternative mechanism of action through which IDH1 mutations may contribute to tumorigenesis. In glioma, the presence of IDH1 mutation was associated with CpG hypermethylation of binding sites for the insulator protein CTCF (CCCTC-binding factor), disabling CTCF binding and correct establishment of DNA loops. As a result, 3D conformational changes of the DNA-architecture affect long-range interactions and subsequently deregulate gene expression [12]. Dysregulation of long-range DNA interactions was recently shown to be involved in leukemogenesis [13, 14]. However, the activation of defined oncogenes by hypermethylation of CTCF sites in the context of IDH1-mutated AML has not been explored. A better understanding of these mechanisms will potentially unravel novel therapeutic targets in the treatment of AML with IDH1 mutation, contributing to overcoming resistance and improving patient outcome.
The aim of this study was to investigate the impact of the IDH1 p.R132H mutation by CTCF site hypermethylation, disruption of 3D DNA conformation and subsequent gene expression changes in AML. Differential gene expression was analyzed in public AML RNA-Seq [15, 16] and microarray data [17] in order to identify deregulated cancer-associated genes in patients with IDH1 mutation. Differential expression results were then integrated with previously generated DNA methylation data [5] and public ChIP-Seq as well as CHIA-PET data in order to correlate upregulated oncogenes with CTCF hypermethylation in the context of altered DNA loop formation. The results were then used for exploration of novel therapeutic options for specific treatment of patients with IDH1-mut AML.
Methods
Cell culture
KG1a myeloid leukemia cells contain a FGFR1OP2-FGFR1 gene fusion but lack other known driver mutations or genetic alterations and were therefore selected as cellular model. Cells were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany), maintained at 0.2–2.0 million cells/ml in RPMI-1640 culture medium (Gibco, Billings, MT, United States) with 20% fetal bovine serum and checked regularly for mycoplasma.
CRISPR base editing and CRISPR knockout
To generate an IDH1 point mutation in KG1a cells, we applied CRISPR base editing [18]. For CRISPR knockout of PDGFRA in IDH1-mut cell clones and knockout of the PDGFRA-CTCF-anchor site in IDH1-wt clones, synthetic sgRNAs were purchased from Synthego (CA, USA) and electroporated together with Cas9 RNP. A detailed description can be found in the Supplementary Methods section. Sanger sequencing primers and sgRNAs are listed in Supplementary Table 1.
RNA, cDNA, RT-qPCR
A detailed description can be found in the Supplementary Methods section. Relative gene expression was calculated using the ΔΔCt method according to the MIQE guidelines [19].
Differential gene expression and survival analysis
Differential gene expression was analyzed in R v4.0.3 with VST values generated from RNA-Seq raw counts using DESeq2 [20]. A detailed description of all used datasets for differential expression and survival analysis can be found in Supplementary Methods section.
DNA methylation and 3D DNA-conformation data
DNA methylation of patients with IDH1-mut and IDH1-wt AML from the Study Alliance Leukemia (SAL) elderly AML cohort (n = 79) was assessed using the Infinium® HumanMethylation450 BeadChip platform and combined with the TCGA AML cohort (n = 194) for comprehensive methylation analysis as described previously [5]. Methylation data are accessible through NCBI’s Gene Expression Omnibus GSE86409. A detailed description can be found in Supplementary Methods section.
Methylation-sensitive restriction digest
A detailed description can be found in the Supplementary Methods section.
Assessment of CTCF binding using ChIP-qPCR
A detailed description can be found in the Supplementary Methods section.
In vitro drug sensitivity screenings, 2-HG detection and treatment
A detailed description can be found in the Supplementary Methods section.
In vivo assessment of dasatinib sensitivity
In vivo experiments were performed using busulfan-pretreated NSG mice [21,22,23,24,25,26] based on the ethical approval for animal studies TVA-V 242—42134/2021 (53-7/21) from the Ministerium für Energiewende, Landwirtschaft, Umwelt, Natur und Digitalisierung (MELUND), Schleswig-Holstein, Germany. A detailed description can be found in the Supplementary Methods.
Single-case AML patient data
Peripheral blood from a 54-years old patient with AML enrolled in the Study Alliance Leukemia (SAL) registry was collected for analysis of neutrophil and leukemic blast counts after informed written consent in accordance with the declaration of Helsinki approved by the local institutional review boards. The patient acquired an IDH1 mutation after failure of 4 lines of therapy, including allogeneic hematopoietic stem cell transplantation. IDH1-positive AML was treated with venetoclax/azacytidine, DLI and cytarabine (araC)/mitoxantrone but resulted in refractory disease. Therefore, therapy with dasatinib was initiated as compassionate use while approval of ivosidenib treatment as compassionate use was awaited. An executive summary of the patient history is provided in Supplementary Table 2.
Statistical analysis
All experiments were performed at minimum in triplicate. Graphs were generated in Microsoft Excel 2016. Error bars represent the standard deviation of the mean. Statistical analysis was performed in R v4.0.3. (R Development Core team, 2013). Data were checked for normal distribution and two-sided Students t-test or ANOVA was performed when normally distributed whereas Wilcoxon or Mann–Whitney U test was used when not normally distributed. The significance level was 0.05.
Results
Upregulation of receptor tyrosine kinase PDGFRA in IDH1-mut AML
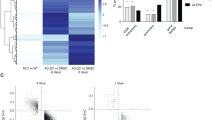
In order to identify upregulated genes in IDH1-mut AML that contribute to IDH1-driven leukemogenesis, we analyzed differential gene expression between patients with IDH1-wt (n = 153) and IDH1-mut (n = 28) AML with hotspot R132 mutation within the BeatAML RNA-Seq dataset [15]. Differential expression analysis identified a total of 132 upregulated genes (log2FoldChange > 1, p-adjusted < 0.05) in patients with IDH1-mut compared to IDH1-wt AML (Supplementary Table 4). Screening of these upregulated genes for classified cancer-associated genes according to the COSMIC Cancer Gene Census (CGC), the OncoKB and the Network of Cancer Genes databases (NCG 7.0) revealed the receptor tyrosine kinase PDGFRA (platelet-derived growth factor alpha) as the most upregulated oncogene in IDH1-mut AML (Fig. 1A). In contrast, PDGFRA expression was not significantly upregulated in IDH2-mut AML (Supplementary Fig. S1A). Gene set enrichment analysis for GO Molecular Function among upregulated genes in patients with IDH1-mut AML revealed enrichment of receptor tyrosine kinase (RTK) activity as well as growth factor binding (Fig. 1B), while no functional categories were significantly enriched among upregulated genes in IDH2-mut AML. Significant upregulation of PDGFRA in IDH1-mut AML was further confirmed using an independent AML gene expression microarray dataset published by Verhaak et al. (Supplementary Fig. S1B) [17]. In order to determine clinical relevance of upregulated PDGFRA expression, we compared overall survival of patients with IDH1-mut AML with high (above median) or low (below median) PDGFRA expression (n = 8 versus n = 8) as well as IDH1-wt patient with high or low PDGFRA (n = 68 versus n = 68) using published clinical data by Bamopoulos et al. [16]. Survival analysis revealed inferior survival of patients with high PDGFRA expression among IDH1-mut but not IDH1-wt groups (Fig. 1C, p = 0.032 vs. p = 0.91). In contrast, no survival difference with respect to PDGFRA expression was found in IDH2-mut versus IDH2-wt groups (Supplementary Fig. S2A). This was further confirmed using the BeatAML clinical data [15] (Supplementary Fig. S2B).
A Differential gene expression in IDH1-mut vs. IDH1-wt AML (data: BeatAML [15]. IDH1-mut patients: n = 28. IDH1-wt patients: n = 153). Blue dots = Differentially expressed genes between IDH1-wt and IDH1-mut AML with cut-off of p.adjusted < 0.05 and log2foldchange > 1. Labeled genes = upregulated cancer-associated genes according to the COSMIC Cancer Gene Census (CGC), the OncoKB and the Network of Cancer Genes databases (NCG 7.0). B Enrichment of cellular interaction-related functional gene groups in IDH1-mut upregulated genes. Gene Ontology enrichment analysis (ShinyGO v0.75) was performed using GO Molecular Function with an FDR cut-off of 0.05. Fold enrichment of GO groups enriched among IDH1-mut upregulated genes is plotted with **FDR < 0.01 and *FDR < 0.05. C Overall survival of IDH1-mut AML patients with high/low PDGFRA expression (n = 8 vs. n = 8) versus IDH1-wt patients with high/low PDGFRA (n = 68 vs. n = 68). PDGFRA high: samples with PDGFRA expression above median. PDGFRA low: samples below median. Method: Kaplan-Maier, significance cut-off p < 0.05. Clinical data: Bamopoulos et al. [16].
Increased CpG hypermethylation within PDGFRA-CTCF anchor in IDH1-mut AML
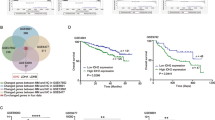
To explore whether upregulation of PDGFRA in IDH1-mut AML may be linked to disrupted gene insulation due to CTCF binding site hypermethylation, we compared DNA methylation of IDH1-mut and IDH1-wt AML samples. Using the combined DNA methylation data from the SAL elderly AML (n = 79) and TCGA cohorts (n = 194), analyzed as described [5], we had previously identified elderly patients with IDH1/2-mut AML as distinct subgroup characterized by a global hypermethylation phenotype. Here, we further identified CpG hypermethylation within a CTCF-anchor region upstream of the PDGFRA gene locus in patients with IDH1-mut AML particularly (Fig. 2A, B). As an in vitro model for confirmation of gene expression data and analysis of DNA methylation and 3D DNA conformation, we generated a mutation knock-in of the IDH1 p.R132H mutation in KG1a AML cells using CRISPR base editing. Successful heterozygous mutation knock-in was validated by sanger sequencing (Supplementary Fig. S3A) and 2-HG production of IDH1-mut clones was confirmed by 2-HG detection assay (Supplementary Fig. S3B). Using quantitative gene expression analysis (RT-qPCR), we confirmed significantly increased DNA methylation of the identified CTCF binding site in IDH1-mut cells compared to IDH1-wt cells (Fig. 2C). We further confirmed upregulated PDGFRA expression in IDH1 p.R132H mutant cells (Fig. 2D). Both increased PDGFRA expression and PDGFRA-CTCF methylation was also observed in naïve KG1a cells treated with exogeneous 2-HG (Fig. 2C, D). We hypothesized that CpG hypermethylation may prevent CTCF binding and subsequently disrupt DNA loop formation and insulation of the PDGFRA gene, leading to PDGFRA upregulation. Therefore, we performed ChIP-qPCR of IDH1-wt and IDH1-mut KG1a cell clones after CTCF antibody pulldown. Quantification of CTCF-occupancy identified decreased CTCF binding by ~60% at the CTCF-anchor region upstream of the PDGFRA locus, suggesting altered DNA loop formation and subsequently disrupted insulation of the PDGFRA gene in IDH1-mut cells (Fig. 2E).
A Top: Visualization of CTCF-anchor site and long-range interaction loops using IGV Genome Browser with ENCODE ChIP-Seq annotation tracks for human CTCF and ENCODE CHIA-PET annotations of CTCF long-range interactions for K562 leukemia cells (hg38) [46]. Methylation tracks (beta-values) of IDH1-wt and IDH1-mut AML patient samples [5] for assessment of CpG methylation within CTCF-anchor site upstream of PDGFRA gene locus (red arrow) using Illumina HM450 data from IDH1-wt (n = 214) and IDH1-mut (n = 33) AML patients [5]. CpG methylation peaks at mentioned CTCF-anchor site (cg18242288) were highlighted in red. B Assessment of CpG methylation within CTCF-anchor site upstream of PDGFRA gene locus using Illumina HM450 data from IDH1-wt (n = 214) and IDH1-mut (n = 33) AML patients [5]. Statistical analysis of beta-logit2 transformed values (M-values) for Illumina HM450 CpG probe cg18242288 using Wilcoxon test with Benjamini Hochberg FDR correction. ****p < 0.0001. C In vitro confirmation of PDGFRA-CTCF hypermethylation based on protection from methylation-sensitive HinP1I digestion of IDH1-wt/-mut KG1a cell clones (2 clones, 3 replicates) and 2-HG treated naïve KG1a cells (3 replicates). Data (mean ± SD) display fold change (FC) in relative expression, Students T-test, *p < 0.05; **p < 0.01; ***p < 0.001. D RT-qPCR confirmation of PDGFRA upregulation in IDH1-mut KG1a cell clones (2 clones, 3 replicates) and 2-HG treated naïve KG1a cells (3 replicates). Data display fold change (FC) in relative expression, Students T-test, *p < 0.05; **p < 0.01; ***p < 0.001. E Decreased CTCF occupancy of PDGFRA-CTCF-anchor region in IDH1-mut KG1a cell clones (2 clones, 3 replicates). Data represent mean ± SD. Students T-test, **p < 0.01.
Drug sensitivity and PDGFRA expression in treated IDH1-mut AML cells
To assess specific drug sensitivity of IDH1-mut AML cells in vitro, IDH1-mut and IDH1-wt KG1a cell clones were treated with the IDH1-mut specific inhibitor ivosidenib, the BCL2-inhibitor venetoclax, the standard chemotherapeutic agent cytarabine (araC) and the demethylating agent 5-azacytidine. Cell viability was assessed using WST assay. Mono-treatment with venetoclax and araC treatment reduced cell viability of both IDH1-mut and IDH1-wt cell clones (Fig. 3A). However, co-treatment with the ABCB1/MDR1/P-GP efflux pump inhibitor verapamil led to increased response of IDH1-mut cells to venetoclax compared to IDH1-wt cells (Supplementary Fig. S4), suggesting that blockade of the ABCB1/MDR1/P-GP efflux pump may be specifically relevant for the clinically benefit of Venetoclax treatment in IDH1-mut AML.
A Normalized cell viability (%) of IDH1-wt and IDH1-mut KG1a clones upon treatment with ivosidenib, dasatinib, venetoclax and araC (72 h) (2 clones, 3 replicates). Data represent mean ± SD. Student´s T-test, significance: *<0.05; **<0.01; ***<0.001. B Normalized cell viability (%) of 2-HG treated naïve KG1a cells, IDH1-mut/PDGFRA-KO cells and IDH1-wt/PDGFRA-CTCF-KO cells upon dasatinib treatment (72 h). Data represent mean ± SD. 2 clones, 3 replicates, Students T-test *p < 0.05; **p < 0.01. C Spleen volume, peripheral blood and bone marrow blast count (assessed by FACS for human CD45+ cells) in NSG mice injected with IDH1-wt versus IDH1-mut KG1a cells with daily dasatinib versus placebo treatment (mean ± SD). n = 5 per group, Mann–Whitney U test *p < 0.05; **p < 0.01.
Confirming clinical data and the specificity of our cellular model, ivosidenib and 5-azacytidine treatment significantly reduced viability of IDH1-mut cells specifically and further resulted in decreased methylation of the PDGFRA-CTCF-anchor site as well as downregulation of PDGFRA gene expression (Fig. 3A, Supplementary Figs. S5, 6). This supports the functional mechanism of ivosidenib and 5-azacytidine in reducing 2-HG mediated DNA hypermethylation, resulting in restored DNA loop formation and subsequent gene expression. However, ivosidenib-mediated downregulation of PDGFRA gene expression was not durable and increased again after ivosidenib treatment pause in vitro (Supplementary Fig. S7).
Increased sensitivity of IDH1-mut AML cells to tyrosine kinase inhibitor dasatinib
Ivosidenib resistance presents a major drawback and 5-azacytidine mediated DNA demethylation acts on a global and rather unspecific scale [27]. Therefore, we explored whether targeting PDGFRA with tyrosine kinase inhibitor (TKI) dasatinib may present an additional treatment option for IDH1-mut AML. IDH1-mut cell clones displayed increased sensitivity towards dasatinib compared to IDH1-wt clones, which was comparable to sensitivity towards IDH1-mut specific inhibitor ivosidenib (Fig. 3A). Cell viability data upon dasatinib and ivosidenib treatment were confirmed using FACS-based cell counting after drug treatment as an independent experimental approach (Supplementary Fig. S8). Confirming correlation of dasatinib sensitivity to upregulated PDGFRA expression, CRISPR knockout of PDGFRA reduced dasatinib sensitivity in IDH1-mut cell clones while CRISPR knockout of the PDGFRA-CTCF site and exogenous 2-HG treatment led to increased PDGFRA expression and dasatinib sensitivity in IDH1-wt cell clones and naïve KG1a cells (Fig. 3B, Supplementary Fig. S9A, B). As dasatinib also inhibits SRC kinases, we also assessed response to crenolanib as TKI without SRC inhibitory activity and observed a similarly increased response of IDH1-mut cell clones as to dasatinib in vitro (Supplementary Fig. S9C), supporting TKI treatment as potential novel treatment option.
Additive effect of dasatinib in combination with ivosidenib
In vitro combination of ivosidenib and dasatinib resulted in an additive combination effect on IDH1-mut cells already at low ivosidenib concentrations, whereas in IDH1-wt cells combinations only appeared to be beneficial at the highest ivosidenib concentrations (Supplementary Fig. S10). To assess whether previous treatment with ivosidenib or 5-azacytidine may affect response to dasatinib in vitro, we treated IDH1-wt and IDH1-mut cell clones sequentially with ivosidenib and dasatinib or 5-azacytidine and dasatinib. Both pre-treatments did not reduce sensitivity of IDH1-mut cells to dasatinib (Supplementary Fig. S11), suggesting that the beneficial effect of dasatinib is not abrogated by previous ivosidenib or 5-azacytidine treatment. These data suggest dasatinib as potential candidate for specific treatment of patients with IDH1-mut AML.
Increased dasatinib sensitivity in vivo and in a patient with IDH1-mut AML
To investigate dasatinib sensitivity in vivo, we generated an in vivo mouse model where IDH1-wt and IDH1-mut KG1a cells were injected into NSG mice that were subsequently treated with dasatinib or placebo. Dasatinib treatment of animals with IDH1-mut KG1a cells resulted in a significant decrease of spleen volume compared to placebo treatment (Fig. 3C). This treatment effect was not observed for animals with IDH1-wt KG1a cells. Additionally, leukemic blast count in peripheral blood and murine bone marrow were assessed by FACS analysis of human CD45-positive cells and revealed a significant decrease in leukemic burden in dasatinib-treated animals injected with IDH1-mut but not IDH1-wt KG1a cells (Fig. 3C).
In addition, in a single-case study of a patient with refractory AML with IDH1 mutation, dasatinib treatment increased the number of segmented neutrophils while the number of leukemic blasts decreased (Fig. 4). Performance status of the patient improved dramatically and treatment was administered in an out-patient setting. However, response to dasatinib was not durable and refractory relapse was noted after 2 months. Subsequent treatment with Ivosidenib failed to induce meaningful disease control (Fig. 4).
Top: Phase contrast images (40x) after Pappenheim staining, Bottom left: Segmented neutrophil count over time [days] under dasatinib (red arrow) and ivosidenib (black arrow) treatment, day 1 = first day of treatment with dasatinib as start of treatment cycle 4 after 3 previous treatment cycles, Bottom right: Blast count [%] over treatment time [days] under dasatinib and ivosidenib treatment.
Discussion
In this study, we identified receptor tyrosine kinase PDGFRA as upregulated oncogene in AML with IDH1 p.R132H mutation. Furthermore, we provide first evidence that PDGFRA upregulation occurs as a result of disrupted DNA loop formation and subsequent loss of gene insulation due to CpG hypermethylation within a CTCF-anchor region upstream of the PDGFRA gene locus in AML. IDH mutations have previously been associated with global DNA hypermethylation in AML as a result of the generation of the oncometabolite 2-HG, which inhibits TET-mediated DNA demethylation [5, 9]. When located within promoter regions of genes involved in cell differentiation, CpG hypermethylation has been shown to lead to a block of myeloid differentiation [7]. However, a role of 2-HG mediated hypermethylation in oncogene activation due to disrupted DNA architecture has not been shown in the context of IDH1-mut AML so far.
Aberrant 3D DNA architecture has generally been reported for different malignancies when compared to healthy controls [13, 28,29,30,31,32,33]. In the context of developmental disorders, Melo et al. identified complex genomic rearrangements and TAD-shuffling, which was associated with phenotypic changes. In leukemia, Hyle et al. showed that CTCF depleting results in loss of MYC expression by disrupted MYC promotor-enhancer interaction in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) [33]. With regard to epigenetic mutations, changes in 3D architecture and subsequently altered gene expression has been reported as well [31, 34, 35]. In AML, disrupted CTCF binding at the CTCF binding site between the HOXA7 and HOXA9 genes was reported to prevent DNA looping and alter expression of posterior HOXA genes [14]. In IDH1-mutant AML, Wilson et al. recently investigated differentially methylated regions (DMR) with regard to enhancer association and identified high enrichment for enhancers in hypermethylated IDH1-mut specific DMRs [36]. However, an association to differential gene expression of the correlating genes could not be identified. Our data link disrupted 3D DNA architecture in IDH1-mut AML to upregulated oncogene expression, providing novel insights into 3D DNA alterations as additional leukemogenesis-driving mechanism in AML.
Here we show that upregulation of tyrosine kinase PDGFRA is associated with altered 3D DNA conformation in IDH1-mut AML and therefore may represent a novel therapeutic target in IDH1-mut AML. PDGFRA is involved in cell proliferation, migration and invasion, cell survival and chemotaxis [37]. In the Network of Cancer Genes (NGC) project it has also been classified as driver gene with oncogenic potential and its overexpression has been reported in breast cancer, ovarian cancer and thyroid cancer [38,39,40]. Yet, activation of PDGFRA expression in connection with altered DNA conformation as potential leukemogenic mechanism in IDH1-mut AML has not been suggested. We provide evidence for decreased CTCF binding at the CTCF-anchor region upstream of PDGFRA due to increased DNA methylation, suggesting disrupted DNA loop formation and insulation and subsequent upregulation of PDGFRA expression. Upregulated PDGFRA-CTCF methylation and PDGFRA expression was also confirmed upon exogenous treatment with 2-HG, confirming 2-HG mediated CTCF hypermethylation as mechanism of action upregulating PDGFRA. This finding is supported by the work from the B. Bernstein research group showing disrupted DNA looping and subsequent activation of PDGFRA expression in IDH1-mut glioma [12]. Our finding of upregulated PDGFRA expression in connection with altered DNA looping in IDH1-mut AML now extends this mechanism of action beyond the context of glioma. As observed in clinical data of IDH1-mut AML patients, IDH1-mut glioma patients with high PDGFRA expression also showed significantly worse survival rates, supporting a strong clinical relevance of this mechanism. These data strengthen the hypothesis that the correlation between IDH1 mutation-dependent DNA conformational changes and upregulated PDGFRA expression may be a more global mechanism across different cancer types.
Concerning treatment options for IDH1-mut AML, the small-molecule inhibitor ivosidenib presents an effective therapeutic agent, leading to remissions in IDH1-mut AML patients [41]. However, responses are often not durable and relapse and drug resistance present a major problem associated with ivosidenib monotherapy, pointing out the importance of identifying novel treatment strategies [11]. For this purpose, the identification of tyrosine kinase PDGFRA as novel target may not only be of biological or prognostic [42] but also of therapeutic relevance.
Here, we provide evidence that treatment with tyrosine kinase inhibitor dasatinib may present a novel treatment option for AML patients with IDH1 p.R132H mutation. While PDGFRA knockout reduced dasatinib sensitivity in IDH1-mut clones, knockout of the PDGFRA-CTCF site and exogenous 2-HG treatment increased dasatinib sensitivity in IDH1-wt clones and naïve KG1a cells, confirming the specific correlation of dasatinib response to PDGFRA expression. The antileukemic effect of dasatinib was confirmed in an in vivo mouse model of IDH1-wt/-mut KG1a and also observed in a single patient with IDH1-mut AML, supporting dasatinib as potential treatment option for IDH1-mut AML. Additionally, our in vitro data also provide a base for the combination of dasatinib and ivosidenib as potential treatment approach.
With regard to combinational therapies, clinical trials have shown promising effects of combination of the demethylating agent 5-azacytidine or BCL2-inhibitor venetoclax with ivosidenib [43, 44]. In our cellular model, the addition of ABCB1/MDR1/P-GP efflux pump inhibitor verapamil led to increased venetoclax response of IDH1-mut cells while venetoclax monotherapy resulted in a similar response of IDH1-mut and IDH1-wt KG1a cells. This suggests that inhibition of the ABCB1/MDR1/P-GP efflux pump may be specifically relevant in this IDH1-mut KG1a model for the observation of mutation-specific sensitivity differences as reported previously [44].
In addition, increased sensitivity of IDH1-mut versus IDH1-wt cells to 5-azacytidine could be confirmed, which supports data from recent clinical trials suggesting 5-azacytidine as combinational partner for ivosidenib treatment of patients with IDH1-mut AML [43]. However, as 5-azacytidine treatment leads to DNA demethylation on a global scale rather than specific demethylation of the PDGFRA-CTCF site [27], Treatment with dasatinib as TKI may add a more targeted treatment option for IDH1-mut AML specifically.
The suggestion of dasatinib as IDH1 mutant-specific treatment is supported by data from Tavor et al. [45] revealing a significant enrichment of samples with IDH1 mutations (33.3%) versus IDH1-wt AML samples (14.3%) among those responding to dasatinib ex vivo (hypergeometric test, p = 0.022). Tavor and colleagues also reported an enrichment of FLT3-ITD and IDH2-mut samples among dasatinib responders in this study. However, our analysis of the underlying RNA-Seq data (BeatAML) [15] revealed no significant difference in expression of PDGFRA in IDH2-mut versus IDH1-wt AML samples (Supplementary Fig. S1A). This suggests that the underlying mechanism of IDH2 mutant-specific sensitivity to dasatinib may differ from our observed IDH1 mutant-specific dasatinib sensitivity and may be related to other dasatinib targets than PDGFRA. This would be compatible with the data from Wilson et al. reporting differences in global hypermethylation patterns of IDH1-mut and IDH2-mut AML [36]. Specific promoter methylation differences may then translate into different downstream gene expression and pathway activation and may therefore provide a possible explanation for a different underlying mechanism of dasatinib response.
IDH1-mut specificity of dasatinib is further supported by the reduced leukemic blast count that was observed in a patient with refractory IDH1-mut AML after dasatinib treatment. As ivosidenib or 5-azacytidine pre-treatment did not abrogate the beneficial effect of dasatinib in IDH1-mut AML cells and as the combination of dasatinib and ivosidenib appeared to be beneficial, our data suggest dasatinib as novel treatment option, also in combinations, for patients with IDH1 p.R132H mutant AML.
In conclusion, our data suggest that IDH1 p.R132H mutation induces changes in the 3D DNA architecture leading to alteration of long-range gene interactions in AML. We show that methylation-dependent disruption of CTCF binding at the CTCF-anchor upstream of PDGFRA leads to upregulated PDGFRA expression. As PDGFRA overexpression correlated with decreased survival of patients with IDH1-mutant AML, we reason that treatment with TKI dasatinib may present a novel therapeutic option for improved treatment for patients with IDH1 p.R132H mutant AML. This may be of particular interest for elderly patients with mutant IDH1, which present a large proportion of patients and show a particularly high IDH mutation frequency and, so far, poor outcome with high resistance and relapse rates.
Data availability
The datasets analyzed during the current study are available at the dbGaP repository with the study ID is 30641 and accession ID is phs001657 or at the NCBI Gene Expression Omnibus repository (GSE146173, GSE6891, GSE86409). All further data generated during this study are included in this published article and its Supplementary Information files.
References
Montalban-Bravo G, DiNardo CD. The role of IDH mutations in acute myeloid leukemia. Future Oncol. 2018;14:979–93.
Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 2019;36:70–87.
Bullinger L, Döhner K, Döhner H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J Clin Oncol. 2017;35:934–46.
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47.
Silva P, Neumann M, Schroeder MP, Vosberg S, Schlee C, Isaakidis K, et al. Acute myeloid leukemia in the elderly is characterized by a distinct genetic and epigenetic landscape. Leukemia. 2017;31:1640–4.
Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017;31:272–81.
Al-Khallaf H. Isocitrate dehydrogenases in physiology and cancer: biochemical and molecular insight. Cell Biosci. 2017;7:37.
Molenaar RJ, Maciejewski JP, Wilmink JW, van Noorden CJF. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene. 2018;37:1949–60.
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67.
Golub D, Iyengar N, Dogra S, Wong T, Bready D, Tang K, et al. Mutant Isocitrate Dehydrogenase Inhibitors as Targeted Cancer Therapeutics. Front Oncol. 2019;9:417.
Donker ML, Ossenkoppele GJ. Evaluating ivosidenib for the treatment of acute myeloid leukemia. Expert Opin Pharmacother. 2020;21:2205–13.
Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2015;529:110–4.
Yang L, Chen F, Zhu H, Chen Y, Dong B, Shi M, et al. 3D genome alterations associated with dysregulated HOXA13 expression in high-risk T-lineage acute lymphoblastic leukemia. Nat Commun. 2021;12:3708.
Luo H, Wang F, Zha J, Li H, Yan B, Du Q, et al. CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood. 2018;132:837–48.
Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31.
Bamopoulos SA, Batcha AMN, Jurinovic V, Rothenberg-Thurley M, Janke H, Ksienzyk B, et al. Clinical presentation and differential splicing of SRSF2, U2AF1 and SF3B1 mutations in patients with acute myeloid leukemia. Leukemia. 2020;34:2621–34.
Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–4.
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–4.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Peake K, Manning J, Lewis CA, Barr C, Rossi F, Krieger C. Busulfan as a myelosuppressive agent for generating stable high-level bone marrow chimerism in mice. J Vis Exp. 2015;e52553.
Saland E, Boutzen H, Castellano R, Pouyet L, Griessinger E, Larrue C, et al. A robust and rapid xenograft model to assess efficacy of chemotherapeutic agents for human acute myeloid leukemia. Blood Cancer J. 2015;5:e297.
Wilkinson FL, Sergijenko A, Langford-Smith KJ, Malinowska M, Wynn RF, Bigger BW. Busulfan conditioning enhances engraftment of hematopoietic donor-derived cells in the brain compared with irradiation. Mol Ther. 2013;21:868–76.
Schewe DM, Lenk L, Vogiatzi F, Winterberg D, Rademacher AV, Buchmann S, et al. Larotrectinib in TRK fusion-positive pediatric B-cell acute lymphoblastic leukemia. Blood Adv. 2019;3:3499–502.
Schewe DM, Alsadeq A, Sattler C, Lenk L, Vogiatzi F, Cario G, et al. An Fc-engineered CD19 antibody eradicates MRD in patient-derived MLL-rearranged acute lymphoblastic leukemia xenografts. Blood. 2017;130:1543–52.
Lenk L, Winterberg D, Vogiatzi F, Laqua A, Spory L, Mayar A, et al. Preclinical Evidence for the Efficacy of CD79b Immunotherapy in B-cell Precursor Acute Lymphoblastic. Leuk Hemasphere. 2022;6:e754.
Grövdal M, Karimi M, Tobiasson M, Reinius L, Jansson M, Ekwall K, et al. Azacitidine induces profound genome-wide hypomethylation in primary myelodysplastic bone marrow cultures but may also reduce histone acetylation. Leukemia. 2014;28:411–3.
Melo US, Schöpflin R, Acuna-Hidalgo R, Mensah MA, Fischer-Zirnsak B, Holtgrewe M, et al. Hi-C Identifies Complex Genomic Rearrangements and TAD-Shuffling in Developmental Diseases. Am J Hum Genet. 2020;106:872–84.
Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–8.
Grimmer MR, Costello JF. Cancer: Oncogene brought into the loop. Nature. 2016;529:34–5.
Zhang X, Jeong M, Huang X, Wang XQ, Wang X, Zhou W, et al. Large DNA Methylation Nadirs Anchor Chromatin Loops Maintaining Hematopoietic Stem Cell Identity. Mol Cell. 2020;78:506–21.e.
Flavahan WA, Drier Y, Johnstone SE, Hemming ML, Tarjan DR, Hegazi E, et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature. 2019;575:229–33.
Hyle J, Zhang Y, Wright S, Xu B, Shao Y, Easton J, et al. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer–promoter looping. Nucleic Acids Res. 2019;47:6699–713.
Ghasemi R, Struthers H, Wilson ER, Spencer DH. Contribution of CTCF binding to transcriptional activity at the HOXA locus in NPM1-mutant AML cells. Leukemia. 2021;35:404–16.
Donaldson-Collier MC, Sungalee S, Zufferey M, Tavernari D, Katanayeva N, Battistello E, et al. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat Genet. 2019;51:517–28.
Wilson ER, Helton NM, Heath SE, Fulton RS, Payton JE, Welch JS, et al. Focal disruption of DNA methylation dynamics at enhancers in IDH-mutant AML cells. Leukemia. 2021;36:935–45.
Ding H, Wu X, Boström H, Kim I, Wong N, Tsoi B, et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nat Genet. 2004;36:1111–6.
Li Q, Li M, Zheng K, Tang S, Ma S. Expression pattern analysis and drug differential sensitivity of cancer-associated fibroblasts in triple-negative breast cancer. Transl Oncol. 2021;14:100891.
Lopez-Campistrous A, Adewuyi EE, Williams DC, McMullen TPW. Gene expression profile of epithelial-mesenchymal transition mediators in papillary thyroid cancer. Endocrine. 2020;72:452-61.
Zhang T, Zhang L, Li F Integrative network analysis identifies potential targets and drugs for ovarian cancer. BMC Med Genomics. 2020;13:132.
Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J, et al. Discovery of AG-120 (Ivosidenib): A First-in-Class Mutant IDH1 Inhibitor for the Treatment of IDH1 Mutant Cancers. ACS Med Chem Lett. 2018;9:300–5.
Wang F, Morita K, DiNardo CD, Furudate K, Tanaka T, Yan Y, et al. Leukemia stemness and co-occurring mutations drive resistance to IDH inhibitors in acute myeloid leukemia. Nat Commun. 2021;12:2607.
DiNardo CD, Stein AS, Stein EM, Fathi AT, Frankfurt O, Schuh AC, et al. Mutant Isocitrate Dehydrogenase 1 Inhibitor Ivosidenib in Combination With Azacitidine for Newly Diagnosed Acute Myeloid Leukemia. J Clin Oncol. 2021;39:57–65.
DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 2020;135:791–803.
Tavor S, Shalit T, Ilani NC, Moskovitz Y, Livnat N, Groner Y, et al. Dasatinib response in acute myeloid leukemia is correlated with FLT3/ITD, PTPN11 mutations and a unique gene expression signature. Haematologica. 2020;105:2795–804.
Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014;159:1665–80.
Acknowledgements
We would like to thank Gabriele Riesen for the great help during the in vivo experiments included in this study. Furthermore, we would like to thank SX (Kennedy Krieger Institute Baltimore) for kindly providing the IDH1 p.R132H sgRNA plasmid for CRISPR base editing. This project was funded by the Deutsche José Carreras Leukämie Stiftung (DJCLS).
Funding
This work was supported by a project grant through the Deutsche José Carreras Leukämie Stiftung (DJCLS). Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
SS conducted the major part of the project´s laboratory work (CRISPR cell model generation, CTCF-occupancy and methylation analysis and drug assays in part) as well as differential expression and methylation analysis and paper writing. PS was involved in the development of the original project idea and conducted initial work on CRISPR cell model generation. LL was involved in planning of the in vivo study and conducted all in vivo experiments included in this study and further contributed to manuscript writing and revision. TB, AH and SH were involved in RNA-Seq data analysis and data visualization. LF provided medical treatment including dasatinib and ivosidenib for the single case of IDH1-mutated refractory AML patient and revised paper. MN and LB contributed to project discussions and further development as well as paper corrections. SL und EH conducted the 2-HG measurement and treatments. KR and MB conducted drug screens and drug treatment of cell clones followed by RT-qPCR for methylation and expression. SX kindly provided the sgRNA plasmid for CRISPR base editing and assisted in manuscript correction. CR contributed as contact person for the patient data from the SAL registry. FV and DMS contributed to planning of the in vivo study and assisted in manuscript revision. MS, VY and KS were involved in 3D DNA confirmation data assessment and interpretation. All work was conducted in the research laboratory of CDB at the University Hospital Schleswig-Holstein in Kiel.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Steinhäuser, S., Silva, P., Lenk, L. et al. Isocitrate dehydrogenase 1 mutation drives leukemogenesis by PDGFRA activation due to insulator disruption in acute myeloid leukemia (AML). Leukemia 37, 134–142 (2023). https://doi.org/10.1038/s41375-022-01751-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01751-6
This article is cited by
-
Synthesis and cytotoxic activity evaluation of novel imidazopyridine carbohydrazide derivatives
BMC Chemistry (2024)
-
The impact of DNA methylation on CTCF-mediated 3D genome organization
Nature Structural & Molecular Biology (2024)