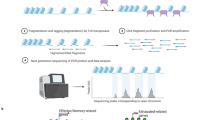
Abstract
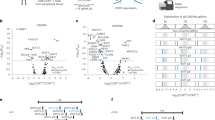
Chimeric antigen receptor T cells (CAR-T) therapy has achieved remarkable therapeutic success in treating a variety of hematopoietic malignancies. However, the high relapse rate and poor in vivo persistence, partially caused by CAR-T cell exhaustion, are still important barriers against CAR-T therapy. It remains largely elusive on the mechanisms of CAR-T exhaustion and how to attenuate exhaustion to achieve better therapeutic efficacy. In this study, we initially observed that CAR-T cells showed rapid differentiation and increased exhaustion after co-culture with tumor cells in vitro, and then performed single-cell ATAC-seq to depict the comprehensive and dynamic landscape of chromatin accessibility of CAR-T cells during tumor cell stimulation. Analyses of differential chromatin accessible regions and motif accessibility revealed that TFs were distinct in each cell type and reconstituted a coordinated regulatory network to drive CAR-T exhaustion. Furthermore, we performed scATAC-seq in patient-derived CAR-T cells and identified BATF and IRF4 as pivotal regulators in CAR-T cell exhaustion. Finally, knockdown of BATF or IRF4 enhanced the killing ability, inhibited exhaustion, and prolonged the persistence of CAR-T cells in vivo. Together, our study unraveled the epigenetic regulatory mechanisms of CAR-T exhaustion and provided new insights into CAR-T engineering to achieve better clinical treatment benefits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
June CH, Sadelain M. Chimeric antigen receptor therapy. N Engl J Med. 2018;379:64–73.
Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med. 2015;21:581–90.
Lynn RC, Weber EW, Sotillo E, Gennert D, Xu P, Good Z, et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature. 2019;576:293–300.
Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. Defining ‘T cell exhaustion’. Nat Rev Immunol. 2019;19:665–74.
Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–13.
Finney OC, Brakke HM, Rawlings-Rhea S, Hicks R, Doolittle D, Lopez M, et al. CD19 CAR T cell product and disease attributes predict leukemia remission durability. J Clin Investig. 2019;129:2123–32.
Henning AN, Roychoudhuri R, Restifo NP. Epigenetic control of CD8(+) T cell differentiation. Nat Rev Immunol. 2018;18:340–56.
Gennert DG, Lynn RC, Granja JM, Weber EW, Mumbach MR, Zhao Y, et al. Dynamic chromatin regulatory landscape of human CAR T cell exhaustion. Proc Natl Acad Sci USA. 2021;30:e2104758118.
Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–9.
Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, et al. Transcription factor IRF4 Promotes CD8(+) T cell exhaustion and limits the development of memory-like T cells during chronic infection. Immunity. 2017;47:1129–41.e1125.
Liu X, Wang Y, Lu H, Li J, Yan X, Xiao M, et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature. 2019;567:525–9.
Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature. 2019;571:211–8.
Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37:925–36.
Sheih A, Voillet V, Hanafi LA, DeBerg HA, Yajima M, Hawkins R, et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat Commun. 2020;11:219.
Deng Q, Han G, Puebla-Osorio N, Ma MCJ, Strati P, Chasen B, et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat Med. 2020;26:1878–87.
Wang W, Fasolino M, Cattau B, Goldman N, Kong W, Frederick MA, et al. Joint profiling of chromatin accessibility and CAR-T integration site analysis at population and single-cell levels. Proc Natl Acad Sci USA. 2020;117:5442–52.
Chen GM, Chen C, Das RK, Gao P, Chen CH, Bandyopadhyay S, et al. Integrative bulk and single-cell profiling of premanufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov. 2021;11:2186–99.
Zhang H, Hu Y, Shao M, Teng X, Jiang P, Wang X, et al. Dasatinib enhances anti-leukemia efficacy of chimeric antigen receptor T cells by inhibiting cell differentiation and exhaustion. J Hematol Oncol. 2021;14:113.
Yang Y, Kohler ME, Chien C, Sauter C, Jacoby E, Yan C, et al. TCR engagement negatively affects CD8 but not CD4 CAR T cell expansion and leukemic clearance. Sci Transl Med. 2017;9:eaag1209.
Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol Cell. 2018;71:858–871.e858.
Miedema JR, Kaiser Y, Broos CE, Wijsenbeek MS, Grunewald J, Kool M. Th17-lineage cells in pulmonary sarcoidosis and Lofgren’s syndrome: friend or foe? J Autoimmun. 2018;87:82–96.
Gray SM, Amezquita RA, Guan T, Kleinstein SH, Kaech SM. Polycomb repressive complex 2-mediated chromatin repression guides effector CD8(+) T cell terminal differentiation and loss of multipotency. Immunity. 2017;46:596–608.
Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–8.
Pipkin ME. Runx proteins and transcriptional mechanisms that govern memory CD8 T cell development. Immunol Rev. 2021;300:100–24.
Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–9.
Chen J, Lopez-Moyado IF, Seo H, Lio CJ, Hempleman LJ, Sekiya T, et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 2019;567:530–4.
Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–53.
Roth TL, Puig-Saus C, Yu R, Shifrut E, Carnevale J, Li PJ, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–9.
Jeannet G, Boudousquié C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci USA. 2010;107:9777–82.
Sommermeyer D, Hudecek M, Kosasih PL, Gogishvili T, Maloney DG, Turtle CJ, et al. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30:492–500.
Turtle CJ, Hanafi LA, Berger C, Hudecek M, Pender B, Robinson E, et al. Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci Transl Med. 2016;8:355ra116.
Agnellini P, Wolint P, Rehr M, Cahenzli J, Karrer U, Oxenius A. Impaired NFAT nuclear translocation results in split exhaustion of virus-specific CD8+ T cell functions during chronic viral infection. Proc Natl Acad Sci USA. 2007;104:4565–70.
Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–20.
Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16:1147–51.
Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol. 2013;13:499–509.
Seo H, Gonzalez-Avalos E, Zhang W, Ramchandani P, Yang C, Lio CJ, et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 2021;22:983–95.
Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–102.
Jin C, Li H, Murata T, Sun K, Horikoshi M, Chiu R, et al. JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Mol Cell Biol. 2002;22:4815–26.
Franchina DG, Dostert C, Brenner D. Reactive oxygen species: involvement in T cell signaling and metabolism. Trends Immunol. 2018;39:489–502.
Scharping NE, Rivadeneira DB, Menk AV, Vignali PDA, Ford BR, Rittenhouse NL, et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat Immunol. 2021;22:205–15.
Tanigawa S, Lee CH, Lin CS, Ku CC, Hasegawa H, Qin S, et al. Jun dimerization protein 2 is a critical component of the Nrf2/MafK complex regulating the response to ROS homeostasis. Cell Death Dis. 2013;4:e921.
Igarashi K, Kurosaki T, Roychoudhuri R. BACH transcription factors in innate and adaptive immunity. Nat Rev Immunol. 2017;17:437–50.
Zhang M, Zhou L, Zhao H, Zhang Y, Wei G, Hong R, et al. Risk factors associated with durable progression-free survival in patients with relapsed or refractory multiple myeloma treated with anti-BCMA CAR T-cell therapy. Clin Cancer Res. 2021;27:6384–92.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–902.e1821.
Stuart T, Srivastava A, Madad S, Lareau CA, Satija R. Single-cell chromatin state analysis with Signac. Nat Methods. 2021;18:1333–41.
Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–3.
Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30.
Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 2010;11:237.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Guo X, Chen F, Gao F, Li L, Liu K, You L, et al. CNSA: a data repository for archiving omics data. Database. 2020;2020:baaa055.
Chen FZ, You LJ, Yang F, Wang LN, Guo XQ, Gao F, et al. CNGBdb: China National GeneBank DataBase. Yi Chuan. 2020;42:799–809.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China, Stem Cell and Translation Research (2018YFA0109300), the Zhejiang Province Science Foundation for Distinguished Young Scholars (LR19H080001), the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2020R01006), and the National Natural Science Foundation of China (81870080, 91949115, 82000187, 81900176). Thanks for the technical support by the Core Facilities, Zhejiang University School of Medicine.
Author information
Authors and Affiliations
Contributions
PJ and PQ conceived the project and designed the experiments. PJ analyzed data and wrote the manuscript. ZZ conducted CAR-T cells with BATF and IRF4 knockdown and performed assays in vivo. ZL performed assays in vitro. YXH and XL were involved in patient sample acquisition. YLH, XZ, HZ, MZ, JD, TG provided cell culture and molecular biology assistance. HH and PQ supervised the overall project and co-wrote the manuscript. All authors contributed to reading and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, P., Zhang, Z., Hu, Y. et al. Single-cell ATAC-seq maps the comprehensive and dynamic chromatin accessibility landscape of CAR-T cell dysfunction. Leukemia 36, 2656–2668 (2022). https://doi.org/10.1038/s41375-022-01676-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01676-0
This article is cited by
-
Epigenetic checkpoints regulate the fate and function of CAR-T cells
Nature Immunology (2024)
-
Deciphering and advancing CAR T-cell therapy with single-cell sequencing technologies
Molecular Cancer (2023)
-
Insights gained from single-cell analysis of chimeric antigen receptor T-cell immunotherapy in cancer
Military Medical Research (2023)
-
Advancing CAR T cell therapy through the use of multidimensional omics data
Nature Reviews Clinical Oncology (2023)
-
Immunotherapy in hematologic malignancies: achievements, challenges and future prospects
Signal Transduction and Targeted Therapy (2023)