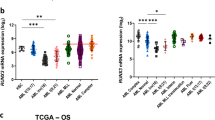
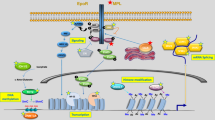
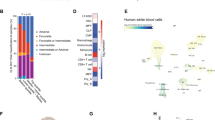
Abstract
RUNX1 is a critical transcription factor for the emergence of definitive hematopoiesis and the precise regulation of adult hematopoiesis. Dysregulation of its regulatory network causes aberrant hematopoiesis. Recurrent genetic alterations in RUNX1, including chromosomal translocations and mutations, have been identified in both inherited and sporadic diseases. Recent genomic studies have revealed a vast mutational landscape surrounding genetic alterations in RUNX1. Accumulating pieces of evidence also indicate the leukemogenic role of wild-type RUNX1 in certain situations. Based on these efforts, part of the molecular mechanisms of disease development as a consequence of dysregulated RUNX1-regulatory networks have become increasingly evident. This review highlights the recent advances in the field of RUNX1 research and discusses the critical roles of RUNX1 in hematopoiesis and the pathobiological function of its alterations in the context of disease, particularly myeloid neoplasms, and clonal hematopoiesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–4.
Bae SC, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, et al. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–14.
Melnikova IN, Crute BE, Wang S, Speck NA. Sequence specificity of the core-binding factor. J Virol. 1993;67:2408–11.
Link KA, Chou FS, Mulloy JC. Core binding factor at the crossroads: determining the fate of the HSC. J Cell Physiol. 2010;222:50–6.
Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070–82.
Bellissimo DC, Speck NA. RUNX1 Mutations in Inherited and Sporadic Leukemia. Front Cell Dev Biol. 2017;5:111.
Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–63.
van Wijnen AJ, Stein GS, Gergen JP, Groner Y, Hiebert SW, Ito Y, et al. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004;23:4209–10.
Ghozi MC, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci USA. 1996;93:1935–40.
Rennert J, Coffman JA, Mushegian AR, Robertson AJ. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol. 2003;3:4.
Levanon D, Glusman G, Bangsow T, Ben-Asher E, Male DA, Avidan N, et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262:23–33.
Challen GA, Goodell MA. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol. 2010;38:403–16.
Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–7.
Imai Y, Kurokawa M, Tanaka K, Friedman AD, Ogawa S, Mitani K, et al. TLE, the human homolog of groucho, interacts with AML1 and acts as a repressor of AML1-induced transactivation. Biochem Biophys Res Commun. 1998;252:582–9.
Draper JE, Sroczynska P, Leong HS, Fadlullah MZH, Miller C, Kouskoff V, et al. Mouse RUNX1C regulates premegakaryocytic/erythroid output and maintains survival of megakaryocyte progenitors. Blood. 2017;130:271–84.
Liu X, Zhang Q, Zhang DE, Zhou C, Xing H, Tian Z, et al. Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis. Leukemia. 2009;23:739–45.
Sakurai H, Harada Y, Ogata Y, Kagiyama Y, Shingai N, Doki N, et al. Overexpression of RUNX1 short isoform has an important role in the development of myelodysplastic/myeloproliferative neoplasms. Blood Adv. 2017;1:1382–6.
Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–30.
Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–9.
Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–97.
Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708.
Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, et al. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–63.
North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–75.
Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–5.
Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–91.
Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304.
Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504.
Lordier L, Bluteau D, Jalil A, Legrand C, Pan J, Rameau P, et al. RUNX1-induced silencing of non-muscle myosin heavy chain IIB contributes to megakaryocyte polyploidization. Nat Commun. 2012;3:717.
Bluteau D, Glembotsky AC, Raimbault A, Balayn N, Gilles L, Rameau P, et al. Dysmegakaryopoiesis of FPD/AML pedigrees with constitutional RUNX1 mutations is linked to myosin II deregulated expression. Blood. 2012;120:2708–18.
Jalagadugula G, Mao G, Kaur G, Goldfinger LE, Dhanasekaran DN, Rao AK. Regulation of platelet myosin light chain (MYL9) by RUNX1: implications for thrombocytopenia and platelet dysfunction in RUNX1 haplodeficiency. Blood. 2010;116:6037–45.
Kuvardina ON, Herglotz J, Kolodziej S, Kohrs N, Herkt S, Wojcik B, et al. RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation. Blood. 2015;125:3570–9.
Huret JL, Ahmad M, Arsaban M, Bernheim A, Cigna J, Desangles F, et al. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013;41:D920–4.
Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31.
Goemans BF, Zwaan CM, Miller M, Zimmermann M, Harlow A, Meshinchi S, et al. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005;19:1536–42.
Shimada A, Taki T, Tabuchi K, Tawa A, Horibe K, Tsuchida M, et al. KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): a study of the Japanese Childhood AML Cooperative Study Group. Blood. 2006;107:1806–9.
Borthakur G, Kantarjian H. Core binding factor acute myelogenous leukemia-2021 treatment algorithm. Blood Cancer J. 2021;11:114.
Prebet T, Boissel N, Reutenauer S, Thomas X, Delaunay J, Cahn JY, et al. Acute myeloid leukemia with translocation (8;21) or inversion (16) in elderly patients treated with conventional chemotherapy: a collaborative study of the French CBF-AML intergroup. J Clin Oncol. 2009;27:4747–53.
Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. Embo J. 1993;12:2715–21.
Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–84.
Hiebert SW, Reed-Inderbitzin EF, Amann J, Irvin B, Durst K, Linggi B. The t(8;21) fusion protein contacts co-repressors and histone deacetylases to repress the transcription of the p14ARF tumor suppressor. Blood Cells Mol Dis. 2003;30:177–83.
Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D, et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell. 2006;9:249–60.
Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M, et al. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–6.
Okuda T, Cai Z, Yang S, Lenny N, Lyu CJ, van Deursen JM, et al. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–43.
Sun XJ, Wang Z, Wang L, Jiang Y, Kost N, Soong TD, et al. A stable transcription factor complex nucleated by oligomeric AML1-ETO controls leukaemogenesis. Nature. 2013;500:93–7.
Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011;333:765–9.
Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, Cauchy P, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 2014;8:1974–88.
Christen F, Hoyer K, Yoshida K, Hou HA, Waldhueter N, Heuser M, et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): an international study on 331 patients. Blood. 2019;133:1140–51.
Rhoades KL, Hetherington CJ, Harakawa N, Yergeau DA, Zhou L, Liu LQ, et al. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood. 2000;96:2108–15.
Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA. 2001;98:10398–403.
Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74.
Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N, et al. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci USA. 2004;101:15184–9.
Cabezas-Wallscheid N, Eichwald V, de Graaf J, Löwer M, Lehr HA, Kreft A, et al. Instruction of haematopoietic lineage choices, evolution of transcriptional landscapes and cancer stem cell hierarchies derived from an AML1-ETO mouse model. EMBO Mol Med. 2013;5:1804–20.
de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H, et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol. 2002;22:5506–17.
Grisolano JL, O’Neal J, Cain J, Tomasson MH. An activated receptor tyrosine kinase, TEL/PDGFbetaR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA. 2003;100:9506–11.
Nishida S, Hosen N, Shirakata T, Kanato K, Yanagihara M, Nakatsuka S, et al. AML1-ETO rapidly induces acute myeloblastic leukemia in cooperation with the Wilms tumor gene, WT1. Blood. 2006;107:3303–12.
Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest. 2005;115:2159–68.
Wang YY, Zhao LJ, Wu CF, Liu P, Shi L, Liang Y, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA. 2011;108:2450–5.
Abdallah MG, Niibori-Nambu A, Morii M, Yokomizo T, Ideue T, Kubota S, et al. RUNX1-ETO (RUNX1-RUNX1T1) induces myeloid leukemia in mice in an age-dependent manner. Leukemia. 2021;35:2983–8.
Maki K, Yamagata T, Asai T, Yamazaki I, Oda H, Hirai H, et al. Dysplastic definitive hematopoiesis in AML1/EVI1 knock-in embryos. Blood. 2005;106:2147–55.
Maki K, Yamagata T, Yamazaki I, Oda H, Mitani K. Development of megakaryoblastic leukaemia in Runx1-Evi1 knock-in chimaeric mouse. Leukemia. 2006;20:1458–60.
Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, et al. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–24.
Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–75.
Harada H, Harada Y, Tanaka H, Kimura A, Inaba T. Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood. 2003;101:673–80.
Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103:2316–24.
Luo X, Feurstein S, Mohan S, Porter CC, Jackson SA, Keel S, et al. ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants. Blood Adv. 2019;3:2962–79.
Wu D, Luo X, Feurstein S, Kesserwan C, Mohan S, Pineda-Alvarez DE, et al. How I curate: applying American Society of Hematology-Clinical Genome Resource Myeloid Malignancy Variant Curation Expert Panel rules for RUNX1 variant curation for germline predisposition to myeloid malignancies. Haematologica. 2020;105:870–87.
Homan CC, King-Smith SL, Lawrence DM, Arts P, Feng J, Andrews J, et al. The RUNX1 Database (RUNX1db): establishment of an expert curated RUNX1 registry and genomics database as a public resource for familial platelet disorder with myeloid malignancy. Haematologica. 2021;106:3004–7.
Decker M, Lammens T, Ferster A, Erlacher M, Yoshimi A, Niemeyer CM, et al. Functional classification of RUNX1 variants in familial platelet disorder with associated myeloid malignancies. Leukemia. 2021;35:3304–8.
Harada Y, Harada H. Molecular pathways mediating MDS/AML with focus on AML1/RUNX1 point mutations. J Cell Physiol. 2009;220:16–20.
Hayashi Y, Harada Y, Huang G, Harada H. Myeloid neoplasms with germ line RUNX1 mutation. Int J Hematol. 2017;106:183–8.
West AH, Godley LA, Churpek JE. Familial myelodysplastic syndrome/acute leukemia syndromes: a review and utility for translational investigations. Ann N Y Acad Sci. 2014;1310:111–8.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Simon L, Spinella JF, Yao CY, Lavallee VP, Boivin I, Boucher G, et al. High frequency of germline RUNX1 mutations in patients with RUNX1-mutated AML. Blood. 2020;135:1882–6.
Feurstein S, Zhang L, DiNardo CD. Accurate germline RUNX1 variant interpretation and its clinical significance. Blood Adv. 2020;4:6199–203.
Ernst MPT, Kavelaars FG, Lowenberg B, Valk PJM, Raaijmakers M. RUNX1 germline variants in RUNX1-mutant AML: how frequent? Blood. 2021;137:1428–31.
Antony-Debré I, Duployez N, Bucci M, Geffroy S, Micol JB, Renneville A, et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia. 2016;30:999–1002.
Brown AL, Arts P, Carmichael CL, Babic M, Dobbins J, Chong CE, et al. RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML. Blood Adv. 2020;4:1131–44.
Yoshimi A, Toya T, Kawazu M, Ueno T, Tsukamoto A, Iizuka H, et al. Recurrent CDC25C mutations drive malignant transformation in FPD/AML. Nat Commun. 2014;5:4770.
Churpek JE, Pyrtel K, Kanchi KL, Shao J, Koboldt D, Miller CA, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126:2484–90.
Haslam K, Langabeer SE, Hayat A, Conneally E, Vandenberghe E. Targeted next-generation sequencing of familial platelet disorder with predisposition to acute myeloid leukaemia. Br J Haematol. 2016;175:161–3.
Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49:204–12.
Ogawa S. Genetics of MDS. Blood. 2019;133:1049–59.
Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–76.
Xiao W, Chan A, Waarts MR, Mishra T, Liu Y, Cai SF, et al. Plasmacytoid dendritic cell expansion defines a distinct subset of RUNX1-mutated acute myeloid leukemia. Blood. 2021;137:1377–91.
Gaidzik VI, Teleanu V, Papaemmanuil E, Weber D, Paschka P, Hahn J, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–8.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–21.
Kanagal-Shamanna R, Montalban-Bravo G, Sasaki K, Darbaniyan F, Jabbour E, Bueso-Ramos C, et al. Only SF3B1 mutation involving K700E independently predicts overall survival in myelodysplastic syndromes. Cancer. 2021;127:3552–65.
Itzykson R, Solary E. An evolutionary perspective on chronic myelomonocytic leukemia. Leukemia. 2013;27:1441–50.
Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–36.
Elena C, Gallì A, Such E, Meggendorfer M, Germing U, Rizzo E, et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood. 2016;128:1408–17.
Duchmann M, Yalniz FF, Sanna A, Sallman D, Coombs CC, Renneville A, et al. Prognostic Role of Gene Mutations in Chronic Myelomonocytic Leukemia Patients Treated With Hypomethylating Agents. EBioMedicine. 2018;31:174–81.
Woo J, Choi DR, Storer BE, Yeung C, Halpern AB, Salit RB, et al. Impact of clinical, cytogenetic, and molecular profiles on long-term survival after transplantation in patients with chronic myelomonocytic leukemia. Haematologica. 2020;105:652–60.
Tsai SC, Shih LY, Liang ST, Huang YJ, Kuo MC, Huang CF, et al. Biological Activities of RUNX1 Mutants Predict Secondary Acute Leukemia Transformation from Chronic Myelomonocytic Leukemia and Myelodysplastic Syndromes. Clin Cancer Res. 2015;21:3541–51.
Gagelmann N, Badbaran A, Beelen DW, Salit RB, Stölzel F, Rautenberg C, et al. A prognostic score including mutation profile and clinical features for patients with CMML undergoing stem cell transplantation. Blood Adv. 2021;5:1760–9.
Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999;340:1330–40.
Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia. 2020;34:966–84.
Zhao LJ, Wang YY, Li G, Ma LY, Xiong SM, Weng XQ, et al. Functional features of RUNX1 mutants in acute transformation of chronic myeloid leukemia and their contribution to inducing murine full-blown leukemia. Blood. 2012;119:2873–82.
Grossmann V, Kohlmann A, Zenger M, Schindela S, Eder C, Weissmann S, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25:557–60.
Branford S, Kim DDH, Apperley JF, Eide CA, Mustjoki S, Ong ST, et al. Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Leukemia. 2019;33:1835–50.
Adnan Awad S, Dufva O, Ianevski A, Ghimire B, Koski J, Maliniemi P, et al. RUNX1 mutations in blast-phase chronic myeloid leukemia associate with distinct phenotypes, transcriptional profiles, and drug responses. Leukemia. 2021;35:1087–99.
Yin CC, Medeiros LJ, Glassman AB, Lin P. t(8;21)(q22;q22) in blast phase of chronic myelogenous leukemia. Am J Clin Pathol. 2004;121:836–42.
Bacher U, Haferlach T, Alpermann T, Zenger M, Hochhaus A, Beelen DW, et al. Subclones with the t(9;22)/BCR-ABL1 rearrangement occur in AML and seem to cooperate with distinct genetic alterations. Br J Haematol. 2011;152:713–20.
Ochi Y, Yoshida K, Huang YJ, Kuo MC, Nannya Y, Sasaki K, et al. Clonal evolution and clinical implications of genetic abnormalities in blastic transformation of chronic myeloid leukaemia. Nat Commun. 2021;12:2833.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98.
Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87.
Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–8.
Bejar R. CHIP, ICUS, CCUS and other four-letter words. Leukemia. 2017;31:1869–71.
Kwok B, Hall JM, Witte JS, Xu Y, Reddy P, Lin K, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126:2355–61.
Jajosky AN, Sadri N, Meyerson HJ, Oduro KA Jr., Kelkar A, Fitzgerald B, et al. Clonal cytopenia of undetermined significance (CCUS) with dysplasia is enriched for MDS-type molecular findings compared to CCUS without dysplasia. Eur J Haematol. 2021;106:500–7.
Malcovati L, Galli A, Travaglino E, Ambaglio I, Rizzo E, Molteni E, et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129:3371–8.
Watanabe-Okochi N, Kitaura J, Ono R, Harada H, Harada Y, Komeno Y, et al. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111:4297–308.
Harada Y, Inoue D, Ding Y, Imagawa J, Doki N, Matsui H, et al. RUNX1/AML1 mutant collaborates with BMI1 overexpression in the development of human and murine myelodysplastic syndromes. Blood. 2013;121:3434–46.
Sashida G, Harada H, Matsui H, Oshima M, Yui M, Harada Y, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun. 2014;5:4177.
Behrens K, Maul K, Tekin N, Kriebitzsch N, Indenbirken D, Prassolov V, et al. RUNX1 cooperates with FLT3-ITD to induce leukemia. J Exp Med. 2017;214:737–52.
Nagase R, Inoue D, Pastore A, Fujino T, Hou HA, Yamasaki N, et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J Exp Med. 2018;215:1729–47.
Hayashi Y, Zhang Y, Yokota A, Yan X, Liu J, Choi K, et al. Pathobiological Pseudohypoxia as a Putative Mechanism Underlying Myelodysplastic Syndromes. Cancer Discov. 2018;8:1438–57.
Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, et al. Combined Cohesin-RUNX1 Deficiency Synergistically Perturbs Chromatin Looping and Causes Myelodysplastic Syndromes. Cancer Discov. 2020;10:836–53.
Aoyagi Y, Hayashi Y, Harada Y, Choi K, Matsunuma N, Sadato D, et al. Mitochondrial Fragmentation Triggers Ineffective Hematopoiesis in Myelodysplastic Syndromes. Cancer Discov. 2022;12:250–69.
Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019;51:27–41.
Barreyro L, Chlon TM, Starczynowski DT. Chronic immune response dysregulation in MDS pathogenesis. Blood. 2018;132:1553–60.
Montalban-Bravo G, Class CA, Ganan-Gomez I, Kanagal-Shamanna R, Sasaki K, Richard-Carpentier G, et al. Transcriptomic analysis implicates necroptosis in disease progression and prognosis in myelodysplastic syndromes. Leukemia. 2020;34:872–81.
Nakagawa M, Shimabe M, Watanabe-Okochi N, Arai S, Yoshimi A, Shinohara A, et al. AML1/RUNX1 functions as a cytoplasmic attenuator of NF-κB signaling in the repression of myeloid tumors. Blood. 2011;118:6626–37.
Tang X, Sun L, Jin X, Chen Y, Zhu H, Liang Y, et al. Runt-Related Transcription Factor 1 Regulates LPS-Induced Acute Lung Injury via NF-κB Signaling. Am J Respir Cell Mol Biol. 2017;57:174–83.
Bellissimo DC, Chen CH, Zhu Q, Bagga S, Lee CT, He B, et al. Runx1 negatively regulates inflammatory cytokine production by neutrophils in response to Toll-like receptor signaling. Blood Adv. 2020;4:1145–58.
Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–20.
Wong WF, Kohu K, Nakamura A, Ebina M, Kikuchi T, Tazawa R, et al. Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J Immunol. 2012;188:5408–20.
Seo W, Shimizu K, Kojo S, Okeke A, Kohwi-Shigematsu T, Fujii SI, et al. Runx-mediated regulation of CCL5 via antagonizing two enhancers influences immune cell function and anti-tumor immunity. Nat Commun. 2020;11:1562.
Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, et al. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2015;17:165–77.
Wu D, Ozaki T, Yoshihara Y, Kubo N, Nakagawara A. Runt-related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 protein in response to DNA damage through complex formation and acetylation. J Biol Chem. 2013;288:1353–64.
Satoh Y, Matsumura I, Tanaka H, Harada H, Harada Y, Matsui K, et al. C-terminal mutation of RUNX1 attenuates the DNA-damage repair response in hematopoietic stem cells. Leukemia. 2012;26:303–11.
Antony-Debre I, Manchev VT, Balayn N, Bluteau D, Tomowiak C, Legrand C, et al. Level of RUNX1 activity is critical for leukemic predisposition but not for thrombocytopenia. Blood. 2015;125:930–40.
Esposito MT, Zhao L, Fung TK, Rane JK, Wilson A, Martin N, et al. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat Med. 2015;21:1481–90.
Hayashi Y, Yokota A, Harada H, Huang G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1alpha in cancer. Cancer Sci. 2019;110:1510–7.
Lee SH, Che X, Jeong JH, Choi JY, Lee YJ, Lee YH, et al. Runx2 protein stabilizes hypoxia-inducible factor-1α through competition with von Hippel-Lindau protein (pVHL) and stimulates angiogenesis in growth plate hypertrophic chondrocytes. J Biol Chem. 2012;287:14760–71.
Lee SH, Bae SC, Kim KW, Lee YM. RUNX3 inhibits hypoxia-inducible factor-1α protein stability by interacting with prolyl hydroxylases in gastric cancer cells. Oncogene. 2014;33:1458–67.
Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, Littman DR, et al. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood. 2010;115:1610–20.
Muto T, Walker CS, Choi K, Hueneman K, Smith MA, Gul Z, et al. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat Immunol. 2020;21:535–45.
Goyama S, Huang G, Kurokawa M, Mulloy JC. Posttranslational modifications of RUNX1 as potential anticancer targets. Oncogene. 2015;34:3483–92.
Krutein MC, Hart MR, Anderson DJ, Jeffery J, Kotini AG, Dai J, et al. Restoring RUNX1 deficiency in RUNX1 familial platelet disorder by inhibiting its degradation. Blood Adv. 2021;5:687–99.
Goyama S, Schibler J, Cunningham L, Zhang Y, Rao Y, Nishimoto N, et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest. 2013;123:3876–88.
Mill CP, Fiskus W, DiNardo CD, Qian Y, Raina K, Rajapakshe K, et al. RUNX1-targeted therapy for AML expressing somatic or germline mutation in RUNX1. Blood. 2019;134:59–73.
Fu L, Fu H, Tian L, Xu K, Hu K, Wang J, et al. High expression of RUNX1 is associated with poorer outcomes in cytogenetically normal acute myeloid leukemia. Oncotarget. 2016;7:15828–39.
Illendula A, Gilmour J, Grembecka J, Tirumala VSS, Boulton A, Kuntimaddi A, et al. Small Molecule Inhibitor of CBFβ-RUNX Binding for RUNX Transcription Factor Driven Cancers. EBioMedicine. 2016;8:117–31.
Ritter M, Klimiankou M, Klimenkova O, Schambach A, Hoffmann D, Schmidt A, et al. Cooperating, congenital neutropenia-associated Csf3r and Runx1 mutations activate pro-inflammatory signaling and inhibit myeloid differentiation of mouse HSPCs. Ann Hematol. 2020;99:2329–38.
Author information
Authors and Affiliations
Contributions
YH and YHar reviewed the literature and wrote the paper; HH edited the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hayashi, Y., Harada, Y. & Harada, H. Myeloid neoplasms and clonal hematopoiesis from the RUNX1 perspective. Leukemia 36, 1203–1214 (2022). https://doi.org/10.1038/s41375-022-01548-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01548-7