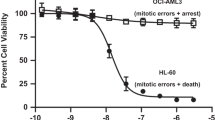
Abstract
Treatment options for patients with relapsed/refractory acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) are scarce. Recurring mutations, such as mutations in isocitrate dehydrogenase-1 and -2 (IDH1/2) are found in subsets of AML and MDS, are therapeutically targeted by mutant enzyme-specific small molecule inhibitors (IDHmi). IDH mutations induce diverse metabolic and epigenetic changes that drive malignant transformation. IDHmi alone are not curative and resistance commonly develops, underscoring the importance of alternate therapeutic options. We were first to report that IDH1/2 mutations induce a homologous recombination (HR) defect, which confers sensitivity to poly (ADP)-ribose polymerase inhibitors (PARPi). Here, we show that the PARPi olaparib is effective against primary patient-derived IDH1/2-mutant AML/ MDS xeno-grafts (PDXs). Olaparib efficiently reduced overall engraftment and leukemia-initiating cell frequency as evident in serial transplantation assays in IDH1/2-mutant but not -wildtype AML/MDS PDXs. Importantly, we show that olaparib is effective in both IDHmi-naïve and -resistant AML PDXs, critical given the high relapse and refractoriness rates to IDHmi. Our pre-clinical studies provide a strong rationale for the translation of PARP inhibition to patients with IDH1/2-mutant AML/ MDS, providing an additional line of therapy for patients who do not respond to or relapse after targeted mutant IDH inhibition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All relevant data are available from the authors upon reasonable request.
References
Showalter MR, Hatakeyama J, Cajka T, VanderVorst K, Carraway KL, Fiehn O, et al. Replication study: the common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Elife. 2017;6:e26030.
Winer ES, Stone RM. Novel therapy in Acute myeloid leukemia (AML): moving toward targeted approaches. Ther Adv Hematol. 2019;10:2040620719860645.
Clark O, Yen K, Mellinghoff IK. Molecular pathways: isocitrate dehydrogenase mutations in cancer. Clin Cancer Res. 2016;22:1837–42.
DiNardo CD, Jabbour E, Ravandi F, Takahashi K, Daver N, Routbort M, et al. IDH1 and IDH2 mutations in myelodysplastic syndromes and role in disease progression. Leukemia. 2016;30:980–4.
Losman JA, Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–52.
Rakheja D, Medeiros LJ, Bevan S, Chen W. The emerging role of d-2-hydroxyglutarate as an oncometabolite in hematolymphoid and central nervous system neoplasms. Front Oncol. 2013;3:169.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30.
Molenaar RJ, Radivoyevitch T, Maciejewski JP, van Noorden CJ, Bleeker FE. The driver and passenger effects of isocitrate dehydrogenase 1 and 2 mutations in oncogenesis and survival prolongation. Biochim Biophys Acta. 2014;1846:326–41.
Wang Y, Wild AT, Turcan S, Wu WH, Sigel C, Klimstra DS, et al. Targeting therapeutic vulnerabilities with PARP inhibition and radiation in IDH-mutant gliomas and cholangiocarcinomas. Sci Adv. 2020;6:eaaz3221.
Norsworthy KJ, Luo L, Hsu V, Gudi R, Dorff SE, Przepiorka D, et al. FDA approval summary: ivosidenib for relapsed or refractory acute myeloid leukemia with an isocitrate dehydrogenase-1 mutation. Clin Cancer Res. 2019;25:3205–9.
Kim ES. Enasidenib: first global approval. Drugs. 2017;77:1705–11.
Liu X, Gong Y. Isocitrate dehydrogenase inhibitors in acute myeloid leukemia. Biomark Res. 2019;7:22.
Upadhyay VA, Brunner AM, Fathi AT. Isocitrate dehydrogenase (IDH) inhibition as treatment of myeloid malignancies: Progress and future directions. Pharm Ther. 2017;177:123–8.
Amatangelo MD, Quek L, Shih A, Stein EM, Roshal M, David MD, et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 2017;130:732–41.
Choe S, Wang H, DiNardo CD, Stein EM, de Botton S, Roboz GJ, et al. Molecular mechanisms mediating relapse following ivosidenib monotherapy in IDH1-mutant relapsed or refractory AML. Blood Adv. 2020;4:1894–905.
Quek L, David MD, Kennedy A, Metzner M, Amatangelo M, Shih A, et al. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat Med. 2018;24:1167–77.
Harding JJ, Lowery MA, Shih AH, Schvartzman JM, Hou S, Famulare C, et al. Isoform switching as a mechanism of acquired resistance to mutant isocitrate dehydrogenase inhibition. Cancer Disco. 2018;8:1540–7.
Intlekofer AM, Shih AH, Wang B, Nazir A, Rustenburg AS, Albanese SK, et al. Acquired resistance to IDH inhibition through trans or cis dimer-interface mutations. Nature. 2018;559:125–9.
Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med. 2017;9:eaal2463.
Sulkowski PL, Oeck S, Dow J, Economos NG, Mirfakhraie L, Liu Y, et al. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature. 2020;582:586–91.
Sulkowski PL, Sundaram RK, Oeck S, Corso CD, Liu Y, Noorbakhsh S, et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat Genet. 2018;50:1086–92.
Molenaar RJ, Radivoyevitch T, Nagata Y, Khurshed M, Przychodzen B, Makishima H, et al. IDH1/2 Mutations sensitize acute myeloid leukemia to PARP inhibition and this is reversed by IDH1/2-mutant inhibitors. Clin Cancer Res. 2018;24:1705–15.
Philip B, Yu DX, Silvis MR, Shin CH, Robinson JP, Robinson GL, et al. Mutant IDH1 promotes glioma formation in vivo. Cell Rep. 2018;23:1553–64.
Song Y, Rongvaux A, Taylor A, Jiang T, Tebaldi T, Balasubramanian K, et al. A highly efficient and faithful MDS patient-derived xenotransplantation model for pre-clinical studies. Nat Commun. 2019;10:366.
Shih AH, Meydan C, Shank K, Garrett-Bakelman FE, Ward PS, Intlekofer AM, et al. Combination targeted therapy to disrupt aberrant oncogenic signaling and reverse epigenetic dysfunction in IDH2- and TET2-mutant acute myeloid leukemia. Cancer Disco. 2017;7:494–505.
Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32:364–72.
Wingett SW, Andrews S. FastQ screen: a tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. Calling somatic SNVs and indels with Mutect2. bioRxiv 2019. https://doi.org/10.1101/861054.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65.
Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–7.
Robinson JT, Thorvaldsdottir H, Wenger AM, Zehir A, Mesirov JP. Variant review with the integrative genomics viewer. Cancer Res. 2017;77:e31–4.
Saito Y, Ellegast JM, Rafiei A, Song Y, Kull D, Heikenwalder M, et al. Peripheral blood CD34(+) cells efficiently engraft human cytokine knock-in mice. Blood. 2016;128:1829–33.
Ellegast JM, Rauch PJ, Kovtonyuk LV, Muller R, Wagner U, Saito Y, et al. inv(16) and NPM1mut AMLs engraft human cytokine knock-in mice. Blood. 2016;128:2130–4.
Lu Y, Kwintkiewicz J, Liu Y, Tech K, Frady LN, Su YT, et al. Chemosensitivity of IDH1-mutated gliomas due to an impairment in PARP1-mediated DNA repair. Cancer Res. 2017;77:1709–18.
Tateishi K, Higuchi F, Miller JJ, Koerner MVA, Lelic N, Shankar GM, et al. The alkylating chemotherapeutic temozolomide induces metabolic stress in IDH1-mutant cancers and potentiates NAD(+) depletion-mediated cytotoxicity. Cancer Res. 2017;77:4102–15.
Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8.
Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7.
Wang HY, Tang K, Liang TY, Zhang WZ, Li JY, Wang W, et al. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J Exp Clin Cancer Res. 2016;35:86.
Stein EM, DiNardo CD, Fathi AT, Pollyea DA, Stone RM, Altman JK, et al. Molecular remission and response patterns in patients with mutant-IDH2 acute myeloid leukemia treated with enasidenib. Blood. 2019;133:676–87.
Madala HR, Punganuru SR, Arutla V, Misra S, Thomas TJ, Srivenugopal KS. Beyond brooding on oncometabolic havoc in IDH-mutant gliomas and AML: current and future therapeutic strategies. Cancers. 2018;10:49.
Rongvaux A, Takizawa H, Strowig T, Willinger T, Eynon EE, Flavell RA, et al. Human hemato-lymphoid system mice: current use and future potential for medicine. Annu Rev Immunol. 2013;31:635–74.
Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–807.
Saito Y, Uchida N, Tanaka S, Suzuki N, Tomizawa-Murasawa M, Sone A, et al. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat Biotechnol. 2010;28:275–80.
Acknowledgements
We thank Yale Pathology Tissue Services especially A. Brooks for research histology services. We thank Yale Animal Resources Center, especially P. Ranney, J. Fonck for animal care. We thank Yale Flow Cytometry Core, especially Lesley Devine, Diane Trotta and Chao Wang for flow cytometry analysis. We thank our patients for donating blood and bone marrow to research. This study was supported in part by the NIH/NIDDK R01DK102792 (to S.H.), The Frederick A. Deluca Foundation and the Vera and Joseph Dresner Foundation (to S.H.), the Howard Hughes Medical Institute (to R.A.F), the General Program of the National Natural Science Foundation of China (Grant No.82170137, to Y.S.), and the Leukemia and Lymphoma Society and the NIH/NCI R01CA266604 (to S.H. and R.S.B.). X.F. was supported by the Young Scientists Fund of the National Natural Science Foundation of China (Grant No. 81801588). Y.G. was supported by the American Society of Hematology Scholar Award. T.T. was supported by AIRC under MFAG 2020 (project ID. 24883). This study was supported by the Animal Modeling Core and the Pilot and Feasibility Program of the Yale Cooperative Center of Excellence in Hematology (NIDDK U54DK106857).
Author information
Authors and Affiliations
Contributions
Conceptualization: SH, RSB, and YS; Methodology: SH, RG, and YS; Investigation: YS, RG, WL, YG, GB, XW, XF, NC, RS, AP, TT, and PM; Data analysis: YS, RG, and SH; Validation: YS, RG, and SH; Writing—original draft: SH, RG, YS; Writing—review & editing: SH, RSB, RG, YS; Funding acquisition: SH and RSB; Resources: RG, AP, AMZ, RAF, and TP; Project administration: SH and RSB; Supervision: SH and RSB.
Corresponding authors
Ethics declarations
Competing interests
Zeidan: AMZ received research funding from Celgene/BMS, Abbvie, Astex, Pfizer, Medimmune/AstraZeneca, Boehringer-Ingelheim, Trovagene/Cardiff Oncology, Incyte, Takeda, Novartis, Aprea, Amgen, and ADC Therapeutics. AMZ participated in advisory boards, had a consultancy with, and/or received honoraria from AbbVie, Otsuka, Pfizer, Celgene/BMS, Jazz, Incyte, Agios, Boehringer-Ingelheim, Novartis, Acceleron, Astellas, Daiichi Sankyo, Cardinal Health, Taiho, Seattle Genetics, BeyondSpring, Trovagene/Cardiff Oncology, Takeda, Ionis, Amgen, Tyme, Aprea, Kura, Janssen, Gilead, and Epizyme. AMZ received travel support for meetings from Pfizer, Novartis, and Trovagene. Flavell: RAF is an advisor to Glaxo Smith Kline, Zai labs and Ventus Therapeutics. Prebet: Boehringer Ingelheim: Research Funding; Novartis: Honoraria; Pfizer: Honoraria; Agios: Consultancy, Research Funding; Jazz Pharmaceuticals: Consultancy, Honoraria, Research Funding; Genentech: Consultancy; Tetraphase: Consultancy; Bristol-Myers Squibb: Honoraria, Research Funding. Bindra: Cybrexa Therapeutics: Consultancy, Equity Ownership. Athena Therapeutics: Equity Ownership. Halene: FORMA Therapeutics: Consultancy.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Gbyli, R., Song, Y., Liu, W. et al. In vivo anti-tumor effect of PARP inhibition in IDH1/2 mutant MDS/AML resistant to targeted inhibitors of mutant IDH1/2. Leukemia 36, 1313–1323 (2022). https://doi.org/10.1038/s41375-022-01536-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01536-x
This article is cited by
-
The diversified role of mitochondria in ferroptosis in cancer
Cell Death & Disease (2023)