Abstract
MIR139 is a tumor suppressor and is commonly silenced in acute myeloid leukemia (AML). However, the tumor-suppressing activities of miR-139 and molecular mechanisms of MIR139-silencing remain largely unknown. Here, we studied the poorly prognostic MLL-AF9 fusion protein-expressing AML. We show that MLL-AF9 expression in hematopoietic precursors caused epigenetic silencing of MIR139, whereas overexpression of MIR139 inhibited in vitro and in vivo AML outgrowth. We identified novel miR-139 targets that mediate the tumor-suppressing activities of miR-139 in MLL-AF9 AML. We revealed that two enhancer regions control MIR139 expression and found that the polycomb repressive complex 2 (PRC2) downstream of MLL-AF9 epigenetically silenced MIR139 in AML. Finally, a genome-wide CRISPR-Cas9 knockout screen revealed RNA Polymerase 2 Subunit M (POLR2M) as a novel MIR139-regulatory factor. Our findings elucidate the molecular control of tumor suppressor MIR139 and reveal a role for POLR2M in the MIR139-silencing mechanism, downstream of MLL-AF9 and PRC2 in AML. In addition, we confirmed these findings in human AML cell lines with different oncogenic aberrations, suggesting that this is a more common oncogenic mechanism in AML. Our results may pave the way for new targeted therapy in AML.
Similar content being viewed by others
Introduction
Mixed-lineage leukemia (MLL) or Lysine Methyltransferase 2A (KMT2A) rearrangements occur in 10% of adult acute leukemias and in 35% of pediatric cases [1, 2]. MLL rearrangements cause in-frame fusions with >60 different genes of which AF4, AF9, AF10, ENL, and ELL are most common [3, 4]. MLL-fusion proteins cause heterogeneous leukemia of lymphoid or myeloid origin and are associated with therapy resistance and poor prognosis [3, 5]. MLL-fusion proteins retain the DNA-binding domain of MLL and deregulate target genes that are normally controlled by MLL [6]. The most common MLL-rearrangement in acute myeloid leukemia (AML) is caused by the t(9;11)(p22q23) fusing MLL to the AF9 gene in-frame and occurs in 30% of MLL-rearranged AMLs [3, 5]. Continuous expression of MLL-AF9 is critical for the survival of AML cells, which suggests that genes deregulated by MLL-AF9 are potential targets for therapy [7, 8].
MicroRNAs (miRNAs) are small non-coding RNAs of 19–22 nt in length and post-transcriptionally regulate mRNAs involved in critical cellular functions [9]. MiRNAs are commonly deregulated in cancer [10, 11]. For instance, the expression of MIR139, encoding miR-139-5p and miR-139-3p, is downregulated and has been identified as a tumor suppressor in AML [12,13,14]. The MIR139 gene is located on chromosome-11 in intron-1 of PDE2A [15]. However, the molecular mechanism of MIR139 silencing in AML is largely unknown. Previously, we have found that miR-139-3p expression is induced by DNA interstrand-crosslinks [12]. There is evidence that miR-139-5p controls the translation machinery through downregulation of Eif4g2 [13, 14].
Polycomb-group (PcG) proteins have been implicated in silencing tumor suppressor genes in various cancers, including MLL-AF9 AML [16,17,18,19]. One class of PcG proteins, polycomb repressive complex 2 (PRC2), consists of the methyltransferase EZH2 or EZH1, EED, SUZ12, and RBBP4 [20]. EZH2 or EZH1 can hypermethylate K27 on Histone-H3, which marks silenced genes [21]. PRC2 contributes to MLL-AF9 pathogenesis and is a target for therapy [22, 23]. However, an additional unknown activity mediated by PRC2 that regulates transcriptional repression, other than blocking the accessibility of chromatin remodeling proteins, cannot be ruled out [24].
Here, we have performed a comprehensive analysis of the tumor suppressor MIR139. We show that MIR139 is a critical tumor suppressor that is silenced by MLL-AF9 in AML. Furthermore, we have identified MIR139 targets that mediate its tumor suppressor activity in AML. Our study has elucidated the molecular silencing mechanism of MIR139 and provides insight into its tumor suppressor activity, offering potential targets of intervention for the poorly prognostic MLL-AF9 AML.
Methods
Generation of knockout mice
C57BL/6 Mir139-deficient mice (Mir139KO) were generated using the CRISPR-Cas9 double-nicking strategy employed by the Cas9D10A mutant and 4 sgRNAs (Mir139KOsgRNA5’plus, Mir139KOsgRNA5’minus, Mir139KOsgRNA3’plus, and Mir139KOsgRNA3’minus) flanking the Mir139 gene as described [25]. Enhancer 1 KO mice (E1KO) and E2KO mice were generated by zygote injection with TrueCut Cas9 Protein v2 (A36499, Thermo Fisher Scientific) and sgRNAs E1KO-5p, E1KO-3p, E2KO-5p, and E2KO-3p (Supplementary Table 1). Heterozygous or homozygous offspring were crossed for at least three generations to reduce CRISPR-Cas9-induced off-target effects. All primers and sgRNAs are listed in Supplementary Table 1. All animal experiments were approved by the Instantie voor Dierenwelzijn (IvD) and the Erasmus Medical Center Animal Welfare and Ethics Committee.
Colony assays, inhibitor experiments, cell culture, and patient samples
For colony assays, 5000 MLL-AF9 cells were plated in MethoCult (M3231, StemCell Technologies), supplemented with IL-3 (1:1000 supernatant), iIL-6 (10 ng/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF) (10 ng/mL), and stem cell factor (SCF) (20 ng/mL), penicillin/streptomycin (100 U/mL) and neomycin 500 µg/mL and expanded for 7 days. DOT1L inhibitor SGC0946 (SML1107, Sigma Aldrich) or UNC1999 (SML0778, Sigma Aldrich) were dissolved in dimethyl sulfoxide (DMSO) and added to the medium at indicated concentrations. Human MLL-AF9 patients were diagnosed according to WHO criteria by cytomorphology, cytogenetics, molecular diagnostics and flow cytometric immunophenotyping. Mononuclear cells were isolated by Ficoll-Paque (density 1.077 g/mL) from peripheral blood or bone marrow. All included patients were positive for the t(9;11)(p22;q23) translocation and samples used for this study contained at least 84% AML blasts as defined by flow cytometry [26]. The use of human MLL-AF9 patient samples was approved by the Erasmus MC Medical Ethics Committee (METC 2016-606). Human MLL-AF9 AML cell lines THP-1, Molm-13, and other AML cell lines MV4-11, HL-60, HEL, and U937 were cultured in Roswell Park Memorial Institute-1640 supplemented with Pen-Strep (100 U/mL), fetal bovine serum (FBS) (10%), and l-Glut (2 mM). AML cell line Kasumi-1 was cultured as above with 20% FBS and AML cell line TF-1 was cultured as above supplemented with human granulocyte-macrophage colony-stimulating factor (5 ng/mL, 300-03-B, Peprotech) and sodium pyruvate (1%). The MLL-AF9 patient samples were cultured in Iscove’s Modified Dulbecco’s Medium, supplemented with FBS (20%), Pen-Strep (100 U/mL), IL-3 (10 ng/mL, 200-03, Peprotech), IL-6 (10 ng/mL, 200-06, Peprotech), TPO (50 ng/mL, 300-18, Peprotech), FLT-3L (50 ng/mL, 300-09, Peprotech), and SCF (50 ng/mL, 300-07, Peprotech).
Statistical analyses
Data are presented as mean ± SEM, unless otherwise specified in the figure legends. Data were compared with the two-tailed independent samples Student’s t test, unless otherwise specified in the figure legends.
Results
MLL-AF9 downregulates Mir139 expression
We generated a murine MLL-AF9 model by transduction of bone marrow (BM) HSPCs with MLL-AF9-expressing retrovirus. MLL-AF9 leukemia cells were positive for c-Kit, GR-1, CD16/CD32, and CD11b and negative for CD3 expression, suggesting a myeloid progenitor origin of the leukemia cells (Fig. 1A). As expected from previous studies [27], DOT1L-inhibitors diminished cell survival of MLL-AF9 cells (Supplementary Fig. 1A). To validate our model, we performed proteomics and observed differential protein expression in MLL-AF9 cells (Supplementary Fig. 1B, Supplementary Table 2). The most upregulated protein was 15-hydroxyprostaglandin dehydrogenase (HPGD), suggesting a role for the HPGD pathway in MLL-AF9 leukemogenesis (Supplementary Fig. 1B). We performed Ingenuity Pathway Analysis of differentially expressed proteins and found that pathways involved in Cell Death and Survival (P < 2.74e−3), Hematological System Development and Function (P < 2.60e−3), and Hematological Diseases (P < 2.60e−3) were deregulated (Supplementary Fig. 1C). Transcriptomics and Gene Set Enrichment Analyses showed enrichment of MLL-AF9 targets [28] in the upregulated fraction of genes in MLL-AF9 cells compared to WT HSPCs (Supplementary Fig. 1D). In contrast, myeloid differentiation factors such as C/ebpa, Csf1r, Csf3r, and Id2 were downregulated (Supplementary Fig. 1E). In concordance with previous studies [22, 29], we found enrichment of PRC1 and PRC2 targets, class-I (EZH2-controlled) and class-II (EZH2-independent) PcG target genes in the set of downregulated genes in MLL-AF9 cells compared to WT HSPCs (Supplementary Fig. 1F).
A Flow cytometric analysis of surface marker expression of MLL-AF9 or MLL-AF9-Mir139KO cells stained for myeloid lineage markers CD11b, CD16/32, c-Kit, GR-1 and lymphoid lineage marker CD3. B Heatmap showing the differential gene expression profiles of WT HSPCs (WT), Mir139KO HSPCs (KO), and MLL-AF9 cells (MA9). C The number of colony-forming units (CFUs) per 5000 empty vector control (−) and MLL-AF9 (MA9) transduced WT and Mir139KO HSPCs are shown. Total number of colonies (50–250 cells/colony) and large colonies (>250 cells/colony) are presented. Micrographs depict representative colonies. Scale bar indicates 100 µm. The two-tailed unpaired student’s t test was used for the statistical analysis. D Expression of miR-139-5p and miR-139-3p in MA9 cells and MLL-AF9-Mir139KO relative to snRNA U6 and EV transduced HSPCs (EV) as determined by miRNA qPCR is shown. The two-tailed unpaired student’s t test with Welch’s correction was used for the statistical analysis. The presented data are representative of two experiments. E Expression of Pde2a in MA9 cells and WT HSPCs in fragments per kilobase and per million mapped fragments (FPKM) as determined by RNA-seq is shown. The Wald’s test with Benjamin–Hochberg correction was used for the statistical analysis. All graphs show mean ± SEM.
MIR139 is downregulated in AML patients including cases with MLL rearrangements and in THP-1 cells that express MLL-AF9 compared with normal HSPCs and myeloid cells including monocytes, macrophages, and neutrophils [13]. We therefore asked whether genomic Mir139 deletion collaborates with MLL-AF9 in AML outgrowth. Mir139KO mice developed normally and had expected HSPC counts and hematopoietic cell types in peripheral blood and BM [25]. Next, we asked whether Mir139 influences genes that have potential effects on pathways other than those that are critical for hematopoiesis. We found that WT and Mir139KO HSPCs were indistinguishable at the transcriptome and proteome level, including Pde2a host gene expression (Fig. 1B, Supplementary Fig. 2A, Supplementary Table 3). These data suggest that MIR139 is expressed at too low levels in normal HSPCs to sufficiently downregulate its targets to detectable levels. In contrast, genomic deletion of Mir139 enhanced primary and secondary colony-forming capacity (more colonies) and proliferation (larger colonies) of MLL-AF9 cells (Fig. 1C). Strikingly, we observed a >60% reduction of miR-139-3p and miR-139-5p expression in MLL-AF9 cells, suggesting that MLL-AF9 silences Mir139 (Fig. 1D). Pde2a expression in MLL-AF9 was unchanged, indicating that Mir139 and the host gene are not co-regulated (Fig. 1E). Together, these results indicate that Mir139 is a target of MLL-AF9.
MiR-139 upregulation and repression of miR-139 targets eliminate MLL-AF9 AML
To determine effects of miR-139 expression on MLL-AF9 cell survival, we generated (DOX)-inducible miR-139-expressing MLL-AF9 clones (MLL-AF9-i139). DOX-treatment of MLL-AF9-i139 cells induced miR-139-5p (up to 130 Fold) and miR-139-3p (up to 100 Fold) expression (Fig. 2A). DOX-treated MLL-AF9-i139 cells lost their colony-forming capacity, whereas DOX-inducible eGFP expressing MLL-AF9 control cells (MLL-AF9-ieGFP) expanded as expected (Fig. 2B). DOX-mediated induction of miR-139 expression caused cell death and apoptosis of MLL-AF9-i139 cells as indicated by Annexin-V positivity and loss of replating capacity (Fig. 2C, D). In addition, induction of miR-139 expression in MLL-AF9-i139 cells cultured in expansion medium without GM-CSF, stimulating mainly leukemia stem cell (LSC) expansion, completely blocked colony outgrowth (Fig. 2D). Similarly, MLL-AF9-i139 cells generated from hematopoietic stem cells (Lin−Sca-1+c-Kit+, LSK)(LSK-MLL-AF9-i139) showed loss of cells upon miR-139 induction (Supplementary Fig. 2B). Together, our results suggest that miR-139 expression also eliminates LSCs.
A Expression levels of miR-139-5p and miR-139-3p in MLL-AF9-i139 clones (n = 3), treated with DOX (5 µg/mL, +) relative to snRNA U6 and mock-treated (−) cells, as determined by miRNA qPCR in triplicate, are shown. The two-tailed unpaired student’s t test was used for the statistical analysis. B The number of colony-forming units (CFU) of MLL-AF9-ieGFP and MLL-AF9-i139 (n = 3) HSPC clones treated with doxycycline (DOX; 5 µg/mL, (+) or mock-treated (−) as control per 8000 cells plated are shown. Data represent the average of three independent experiments. The two-tailed paired student’s t test was used for the statistical analysis. C Representative flow cytometry plot of MLL-AF9-i139 cells with or without DOX stained with propidium iodide and Annexin-V. Data are representative of three independent clones. D Relative number of colony-forming units (CFU) of MLL-AF9-ieGFP and MLL-AF9-i139-1 cells in triplicate, treated with Doxycycline (DOX; 5 µg/mL, (+)) or mock-treated (−) as control per 8000 cells plated are shown of the first plating and the subsequent replating. The cells were cultured with IL-3, IL-6, GM-CSF, and SCF (left panel) or with IL-3, IL-6, and SCF (right panel). The two-tailed paired student’s t test was used for the statistical analysis. E Kaplan–Meier plot showing the survival of mice, transplanted with MLL-AF9-GFP-TetR-KRAB-i139 LSK cells and treated with DOX (4 mg/kg) (red, n = 11) or mock-treated (black, n = 11). The Mantel–Cox test was used for the statistical analysis. F Expression levels of miR-139-5p and miR-139-3p in leukemia cells from mice treated with DOX (n = 4) in E relative to mock-treated and snRNA U6 are shown. Tissues were analyzed from mice when moribund. G Fold change of Eif4g2 (Padj: 0.0002), Hpgd (Padj: 0.0006), and Ptprt (Padj: 0.005) expression in DOX-treated (+DOX) MLL-AF9-i139 clones (n = 3) relative to mock-treated (−DOX) clones (n = 3) as determined by RNA-seq is shown. The Wald’s test with Benjamini–Hochberg correction was used for the statistical analysis. H Boxplots showing the reads (n + 1) of the total six sgRNAs targeting Men1, Dot1l, Mllt3, Eed, Rbbp4, Eif4g2, Hpgd, and Ptprt in MLL-AF9 cells 24 h or 14 days post transduction. Per gene, the order of the sgRNAs in the 24 h sample corresponds to the same sgRNAs in the 14d sample. Blue circles indicate a loss of >5 fold, while red circles indicate a loss of <5 fold of the sgRNA. All graphs depict mean ± SEM.
Next, we asked whether miR-139 expression inhibits MLL-AF9 leukemia development in mice. Mice transplanted with MLL-AF9-i139 cells developed AML with a short latency of 3 weeks. However, DOX-mediated miR-139 induction significantly delayed leukemia development of transplanted mice (Fig. 2E). Mice had malignant MLL-AF9-i139 cells in BM (Supplementary Fig. 2C), blood, and spleen (data not shown). Leukemia cells from mice that were treated with DOX showed an average miR-139-5p and miR-139-3p overexpression induction of eightfold and sevenfold, respectively (Fig. 2F), which suggests that this level of upregulation in escaping MLL-AF9-i139 cells is not sufficient to eliminate MLL-AF9 cells in mice. In full agreement, secondary transplantation experiments with these AML cells show no difference in latency between DOX-treated and non-treated controls (data not shown). These data indicate the tumor-suppressing activities of miR-139 in MLL-AF9 AML.
To gain mechanistic insight, we performed mRNA sequencing of DOX-treated MLL-AF9-i139 and untreated clones and found that a subset of genes were differentially expressed between these populations (Supplementary Fig. 2D, Supplementary Table 4). We established that genes that inhibit cell cycle progressions, such as Btg2, Cdkn1a, and Cdkn2d, and pro-apoptotic genes such as Bmf, Dusp1, Rnasel, Trp53inp1, Ypel3, and Zc3h12a were upregulated in DOX-treated MLL-AF9-i139 cells (Supplementary Table 4, Supplementary Fig. 2F). Furthermore, we noted that predicted miR-139 targets such as Eif4g2, Ptprt and Hpgd were downregulated in miR-139 expressing MLL-AF9 cells (Fig. 2G, Supplementary Table 4).
Next we wanted to validate miR-139 targets that mediate its tumor suppressor activity in MLL-AF9 cells. We performed a genome-wide CRISPR-KO screen to identify genes critical for MLL-AF9 survival or proliferation (Supplementary Fig. 2E). After 14 days, cells with sgRNAs that target such genes will be lost. Confirming the effectivity of our sgRNA screen, we found that sgRNAs targeting Men1 (six sgRNAs), a crucial co-factor for MLL-AF9 [30], the PRC2 subunits Eed (three out of six sgRNAs) and Rbbp4 (five out of six sgRNAs), Dot1l (five out of six sgRNAs) and Mllt3 (four out of six sgRNAs) were lost (Fig. 2H, Supplementary Table 5). We found that MLL-AF9 cells with sgRNAs targeting Ptprt (three out of six) Hpgd (five out of six) and Eif4g2 (five out of six) were also lost, suggesting that expression of our identified miR-139-targets is essential for cell survival (Fig. 2H, Supplementary Table 5). Validation experiments with single sgRNAs targeting Hpgd, Eif4g2, and Ptprt in colony-forming unit assays confirmed these results and showed that the depletion of these miR-139 target genes could, at least in part, phenocopy miR-139 expression in MLL-AF9 cells (Supplementary Fig. 2G). Furthermore, luciferase-based miRNA target validation assays confirmed that Hpgd and, Eif4g2 and Ptprt are direct targets of miR-139 (Supplementary Fig. 2H). Taken together, these data show that miR-139 expression and the downregulation of miR-139 targets inhibit MLL-AF9 leukemia.
Mir139 is epigenetically silenced by PRC2 in MLL-AF9 cells
We next investigated how MIR139 expression is regulated. MIR139 is epigenetically silenced in AML, including the THP-1 cell line that expresses MLL-AF9 [13]. We hypothesized that Mir139 silencing by histone modifications is critical for MLL-AF9 AML. To test this, we treated MLL-AF9 and MLL-AF9-Mir139KO cells with Trichostatin-A (TSA) or valporic acid (VPA), which are inhibitors of class-I and II histone deacetylases. We found that MLL-AF9 cells treated with either TSA or VPA, were significantly reduced in their colony-forming capacity compared with MLL-AF9-Mir139KO cells (Supplementary Fig. 3A). These results suggest a role for MIR139 epigenetic silencing in MLL-AF9 AML expansion and survival.
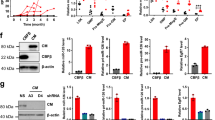
Next, we investigated the epigenetic landscape of Mir139 by reanalyzing publicly available ChIP-seq data sets from mouse MLL-AF9 cells and WT HSPCs [29, 31, 32]. Mir139 is located in intron-1 of the Pde2a host gene with the same orientation, downstream of the highly conserved putative Pde2a promoter (P). As expected, H3K4me3 peaks overlapped with P and H3K27Ac peaks overlapped with enhancer region-1 (E1) and E2 (Fig. 3A). In these genomic regions we noted two POL-II peaks: one located at the first exon of Pde2a and one on E2, in intron-1 of Pde2a (Fig. 3A). We found H3K27me3 levels, which are associated with epigenetic silencing, at P, E1, and E2 in MLL-AF9 cells (Fig. 3A). MLL-AF9 cells treated with UNC1999, a specific inhibitor of the methyltransferases EZH1 and EZH2 [29], showed decreased H3K27me3 and increased H3K27Ac levels compared to UNC2400 control inhibitor-treated cells. In addition, ChIP-seq data revealed SUZ12 binding to P, E1, and E2, which was decreased in cells treated with UNC1999 (Fig. 3A).
A ChIP-seq reads of the Pde2a locus (chr7:108,596,060–108,645,660) are depicted. The top two tracks show H3K4me3 (red) and H3K27Ac (orange) enrichment in common myeloid progenitors (CMPs). The next track indicates RNA Polymerase II (POL-II) binding (purple) of MLL-AF9 cells. The next three tracks depict H3K27me3 (blue), H3K27Ac and SUZ12 (green) enrichment in MLL-AF9 cells treated with UNC2400 (inactive PRC2 inhibitor). The bottom three tracks show the same data of MLL-AF9 cells treated with UNC1999 (active PRC2 inhibitor). Scale bar indicates 5 kilobases (kb). Black bars indicate exons. B The methylation status of CpGs in the PDE2A promoter region, enhancer region 1 (E1) and E2 of THP-1 and Molm-13 cells is depicted. Closed blue circles indicate methylated CpG, open blue circles indicate unmethylated CpG. C Schematic overview of the Pde2a locus (chr7:108,596,060–108,645,660). Fold enrichment of H3K27me3 (orange) and control IgG (white) antibody binding to the indicated regions in MLL-AF9 cells treated with depicted concentrations of UNC1999 as determined by ChIP-qPCR is shown. Arrows indicate the primer sets. The scale bar indicates 2.5 kb. D Viability of WT or Mir139KO MLL-AF9 cells treated with UNC1999 relative to mock-treated WT or Mir139KO MLL-AF9 cells is shown. Data are representative of three experiments. E Expression levels of miR-139-3p and miR-139-5p in MLL-AF9 cells treated with UNC1999 relative to snRNA U6 and untreated MLL-AF9 cells are shown. Data are representative of three experiments. F Viability of Molm-13 cells treated with UNC1999 relative to mock-treated Molm-13 cells, is shown. Depicted data are representative of three experiments. G Expression levels of miR-139-3p and miR-139-5p in Molm-13 cells treated with UNC1999 relative to snRNA U6 and untreated Molm-13 cells are shown. Depicted data are representative of three experiments. H Viability of UNC1999-treated primary MLL-AF9 patient samples (n = 3) relative to untreated cells is shown. I Expression levels of miR-139-5p and miR-139-3p in primary MLL-AF9 patient samples (n = 3) treated with UNC1999 relative to snRNA U6 and untreated cells are shown. All graphs show mean ± SEM. The two-tailed unpaired student’s t test was used in C–E and G. The two-tailed paired student’s t test was used in F, H and I.
Aberrant enhancer DNA methylation is acquired during malignant transformation [33]. PRC2, a chromatin-modifying enzyme that catalyzes histone-H3 lysine 27 trimethylation, is known to have a critical role in MLL-AF9 pathogenesis [22, 23] and prefers binding to methylated DNA via AEBP2 [34]. To address the role of PRC2 in context of MIR139, we determined the CpG methylation status of human MLL-AF9 cell lines Molm-13 and THP-1. We observed that a small percentage of CpGs present in the MIR139 promoter region of Molm-13 (22.73%) and THP-1 (6.36%) are methylated (Fig. 3B). Although there is a lower density of CpGs in the enhancer regions close to MIR139, we detected an increased percentage of methylated CpGs in E1 (67% and 79%) and E2 (60% and 87%) in Molm-13 cells and THP-1 cells, respectively (Fig. 3B).
To experimentally validate the relevance of PRC2-mediated silencing of Mir139, we treated MLL-AF9 cells with UNC1999 and determined cell survival compared with MLL-AF9-Mir139KO cells. UNC1999 activity was confirmed by reduced H3K27me3 levels at the P, E1, and E2 (Fig. 3C). We observed a decreased viability of MLL-AF9 cells compared with MLL-AF9-Mir139KO cells, strongly suggesting that UNC1999 treatment activates miR-139 expression (Fig. 3D). Indeed, loss of cell viability coincided with up to 25-fold increased levels of miR-139-5p and up to 20-fold increased miR-139-3p expression (Fig. 3E). Notably, treatment of MLL-AF9 cells with UNC2400 did not upregulate miR-139 and sustained cell viability (Supplementary Fig. 3B, C). Expression of miR-139-5p was not induced when WT HSPCs were treated with UNC1999 (Supplementary Fig. 3D). Cdkn2a is a known target of PRC2 in MLL-AF9 AML [22, 23]. In agreement, we observed a three to sixfold upregulation of Cdkn2a (Supplementary Fig. 3E). The expression of Pde2a was unaffected by UNC1999 (Supplementary Fig. 3F). This mechanism is conserved between mouse and human, because comparable results were obtained with Molm-13 (Fig. 3F, G), THP-1 (Supplementary Fig. 3G, H) and primary MLL-AF9 patient samples (Fig. 3H, I). In addition, UNC1999 treatment of a panel of AML cell lines caused decreased cell viability and increased miR-139 expression (Supplementary Fig. 3I, J). This suggests that PRC2-mediated MIR139 silencing may not be exclusive for MLL-AF9 AML. Taken together, these data indicate that MLL-AF9 silences MIR139 by a PRC2-mediated mechanism.
E1 and E2 control the expression of Mir139
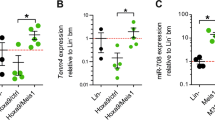
To further investigate the role of the P, E1, and E2 in Mir139 expression, we deleted these genomic regions in mice (Fig. 4A). We were unable to generate mice with genomic deletion of the Pde2a promoter, which overlaps with exon-1, suggesting that a Pde2a KO is embryonically lethal. In contrast, E1 and E2 deletion mutant mice were viable and developed normally. Under physiological conditions, deletion of either E1 or E2 caused a >60% reduction of miR-139-5p expression, a 40–50% reduction of miR-139-3p expression in WT HSPCs and a 70–80% reduction of Pde2a expression (Fig. 4B, C). Next, we investigated whether E1 and E2 are involved in Mir139 silencing in MLL-AF9 cells. Therefore, we generated MLL-AF9-E1KO and MLL-AF9-E2KO cells. Similar to MLL-AF9 cells, these cells were positive for GR-1, CD11b, CD16/32, c-Kit, and negative for CD3 (Fig. 4D). Next, we treated these cells with UNC1999 and noted reduced cell death of MLL-AF9-E1KO and MLL-AF9-E2KO cells compared to MLL-AF9 cells, suggesting that the miR-139 induction is affected (Fig. 4E). Indeed, we found that upon UNC1999 treatment, miR-139-5p and miR-139-3p were not upregulated in MLL-AF9-E1KO cells (Fig. 4F). Notably, Pde2a expression was not affected in UNC1999-treated MLL-AF9-E1KO cells (Fig. 4G). MLL-AF9-E2KO cells showed a significantly reduced miR-139-5p and miR-139-3p induction, which coincided with slightly reduced Pde2a levels with 5 µM UNC1999 (Fig. 4F, G). Together, these data indicate that both E1 and E2 regions control Mir139 expression in AML.
A Schematic representation of CRISPR-Cas9 KO strategy to generate PKO, E1KO, and E2KO mice. Red arrows indicate sgRNAs targeting the promoter region and exon-1 of Pde2a. Green arrows indicate sgRNAs targeting E1 and E2. The first three exons of Pde2a (black bars) are indicated with numbers 1, 2, and 3. The Mir139 encoding sequence is depicted in red. The scale bar indicates 2.5 kb. B Expression levels of miR-139-3p and miR-139-5p in Enhancer 1 KO (E1KO) and E2KO HSPCs relative to snRNA U6 and WT HSPCs are shown C. Expression of Pde2a relative to Gapdh and to WT HSPCs, in E1KO and E2KO HSPCs isolated from three mice per condition is shown. D Flow cytometric analysis of surface marker expression of MLL-AF9-E1KO and MLL-AF9-E2KO cells stained for myeloid lineage markers CD11b, CD16/32, c-Kit, GR-1, and lymphoid lineage marker CD3. E Viability of MLL-AF9 WT, E1KO, and E2KO MLL-AF9 cells treated with indicated concentrations of UNC1999 relative to the mock-treated condition cells is shown. The ANOVA analysis with Bonferroni correction was used for statistical analysis. *In top of graph: comparisons of E1KO and E2KO with WT. Presented data are representative of three independent experiments. F Expression levels of miR-139-3p and miR-139-5p in WT, E1KO, and E2KO MLL-AF9 cells treated with indicated concentrations of UNC1999 relative to snRNA U6 and mock-treated cells are shown. Depicted data are representative of three experiments. G Expression of Pde2a in WT, E1KO, and E2KO MLL-AF9 cells, treated with indicated concentrations of UNC1999 relative to Gapdh and mock-treated cells is shown. Depicted data are representative of three experiments. The two-tailed unpaired student’s t test with Welch’s correction was used for statistical analysis in B–F and G. All graphs indicate mean ± SEM.
POLR2M silences MIR139 in MLL-AF9 cells
To identify novel factors that are involved in transcriptional silencing of Mir139 in MLL-AF9 cells, we performed a genome-wide CRISPR-KO screen [35]. We used MLL-AF9-Mir139KO cells as a reference, which allowed for screening of factors that specifically silence Mir139 transcription. We expected that inactivation of genes encoding for factors responsible for Mir139 silencing would upregulate miR-139 expression and induce apoptosis in MLL-AF9 cells, but not in MLL-AF9-Mir139KO cells (Fig. 5A). We selected all sgRNAs with 10 or more reads in MLL-AF9 cells and MLL-AF9-Mir139KO cells (MLL-AF9-WT + MLL-AF9-Mir139KO ≥ 10) after 14 days of incubation (Fig. 5B). We reasoned that a higher number of sgRNAs lost per gene will increase the chance that these genes are relevant for Mir139 regulation. According to our analysis of control sgRNAs, the chance that a loss of five sgRNAs or more per gene is an artifact is >0.0016%. Out of the genes shown in Fig. 5B, we identified only 16 genes targeted by 5–6 sgRNAs that were significantly reduced by ≥ 10 fold in MLL-AF9 cells compared with MLL-AF9-Mir139KO cells (Supplementary Fig. 4A, Supplementary Table 6). Among these 16 genes, RNA Polymerase 2 Subunit M (Polr2m), involved in stable transcriptional pausing of POL-II [36,37,38], was the only negative transcriptional regulator and was selected for further validation (Fig. 5C). CRISPR-Cas9-mediated KO of Polr2m reduced colony outgrowth and cell viability of MLL-AF9 cells compared with MLL-AF9-139-KO cells (Fig. 5D). In agreement, POLR2M depletion reduced the viability of Molm-13 cells and primary MLL-AF9 cells from patients (Fig. 5E). Furthermore, sgRNA-mediated targeting of POLR2M caused an increase of pri-miR-139 and mature miR-139 expression in Molm-13 cells and in primary MLL-AF9 patient samples compared with control sgRNA treated cells (Fig. 5F, Supplementary Fig. 4B, C). In these experiments, Pde2a expression was not affected by Polr2m depletion (Supplementary Fig. 4D). These results were confirmed in various human AML cell lines with different oncogenic aberrations (Fig. 5G, Supplementary Fig. 4D). Collectively, these data suggest that POLR2M is a silencing factor of MIR139 downstream of PRC2, an oncogenic mechanism that may not be exclusive for MLL-AF9 AML.
A Schematic overview of the sgRNA library screen with MLL-AF9 WT (MA9 WT) and MLL-AF9 Mir139KO (MA9 Mir139KO) cells. B Volcano plot showing –log10 P value (Y axis) and average log2 fold change (X axis) of all targeted genes in MLL-AF9 WT relative to MLL-AF9-Mir139KO 14 days post transduction. The red closed circle indicates Polr2m. Red dashed lines indicate a fold change of ±10. Top 10 of enriched genes are shown. C Boxplots showing the reads (n + 1) of six sgRNAs targeting Polr2m in MA9 WT and MA9 Mir139KO cells. The order of the sgRNAs in the MA9 Mir139KO sample corresponds to the same sgRNAs in the MA9 WT sample. Blue circles indicate a loss of ≥10 fold per sgRNA, while red circles indicate a loss of ≤10 fold of the sgRNA. D MLL-AF9 cells and MLL-AF9 Mir139KO cells were transduced with three viruses expressing sgRNAs targeting Polr2m or three viruses expressing control sgRNAs. The left panel shows the colony-forming units (CFU) of Polr2m-targeted MLL-AF9 cells relative to MLL-AF9 Mir139KO cells. The right panel shows the viability of Polr2m-targeted MLL-AF9 cells relative to MLL-AF9 Mir139KO cells (Y axis). The two-tailed unpaired student’s t test with Welch’s correction was used for statistical analysis. The graphs are representative of three independent experiments. E Viability of POLR2M-targeted Molm-13 cells (left panel) or primary MLL-AF9 patient samples (right panel) relative to control targeted cells. The two-tailed paired student’s t test was used for the statistical analysis. Data are representative of two/three experiments. F Expression levels of pri-miR-139 in POLR2M-targeted Molm-13 cells (left panel) or primary MLL-AF9 patient samples (right panel) relative to control targeted cells. The two-tailed unpaired student’s t test with Welch’s correction was used for statistical analysis. The graphs are representative of two/three independent experiments. G Expression levels of pri-miR-139 in POLR2M-targeted U937, MV4-11, TF-1, or HL-60 cells relative to control targeted cells (left panel). Viability of POLR2M-targeted U937, MV4-11, TF-1, or HL-60 cells relative to control targeted cells (right panel). The two-tailed unpaired student’s t test with Welch’s correction was used for statistical analysis. The graphs are representative of two independent experiments. All graphs are depicted as mean ± SEM.
POLR2M binds to E1, E2, and the transcriptional start site of MIR139
Because POLR2M binds to POL-II to mediate stable transcriptional pausing [36,37,38], we hypothesized that POLR2M is bound at the POL-II interaction sites within the MIR139 locus. We used publicly available data sets to determine enrichment of H3K4me3, H3K27Ac, H3K4me1, and POL-II binding on E1, E2, and P regions (Supplementary Fig. 5). In our analysis, we included a previously identified MIR139 transcriptional start site (TSS) [15] downstream of E2 (Fig. 6A). ChIP-qPCR experiments with chromatin isolated from Molm-13 cells showed significant enrichment of POL-II at P, E1, and E2 regions and the TSS downstream of E2 (Fig. 6B). We observed POLR2M interactions with E1, E2, and the TSS downstream of E2, whereas this was not observed at P (Fig. 6C). Treatment of Molm-13 cells with UNC1999 abrogated POLR2M binding to E1 and E2, but not to the TSS downstream of E2 (Fig. 6D). In UNC1999-treated Molm-13 cells we found strong induction of transcripts originating from the TSS, which was minor at E1 or E2 (Fig. 6E). These data suggest that E1 and E2 in UNC1999-treated cells, strongly induces the transcription by direct interaction with the downstream TSS of MIR139 (Supplementary Fig. 5B). Taken together, these data indicate that POLR2M-binding to E1 and E2 silences the transcription of MIR139 at the TSS downstream of PRC2 in MLL-AF9 AML, which is also illustrated in our model (Fig. 7).
A Schematic representation of the PDE2A locus (chr11:72,605,000–72,644,500) with the indicated promoter (P, yellow) and enhancer (E1 or E2, blue) regions and transcriptional start sites (arrows). The first two exons of PDE2A are depicted with a black horizontal bar. The MIR139 encoding sequence is depicted in red. The location of the primer sets with the respective numbers used for the experiments in this figure is depicted in gray. B, C Fold enrichment of POL-II (yellow) and POLR2M (orange) on the depicted loci relative to IgG (white) as determined by ChIP-qPCR in Molm-13 cells are shown. Data are representative of three experiments. D Same as B and C, except that Molm-13 cells were treated with indicated concentrations of UNC1999 (different shades of orange). Data are representative of two independent experiments. E Schematic overview of the MIR139 locus. The primer sets to detect transcripts coming from E1 (green), E2 (red), and TSS (blue) are shown. The relative expression of transcripts in UNC1999-treated Molm-13-treated cells is shown below. The two-tailed unpaired student’s t test with Welch’s correction was used for statistical analysis in B–E. Data are representative of three experiments. All graphs are depicted as mean ± SEM.
In the top panel, the MIR139 silencing mechanism in MLL-AF9 AML is shown. MLL-AF9 recruits PRC2 to the promoter, E1, E2, and the TSS of the PDE2A locus. POLR2M is recruited to E1, E2, and the TSS. POLR2M silences the transcription of MIR139, while the transcription of the host gene is unaffected. In the bottom panel, the mechanism of transcriptional activation by PRC2 inhibition and POLR2M depletion is shown. POLR2M depletion and PRC2 inhibition by UNC1999, which abrogates POLR2M interaction with the enhancers, results in transcription of MIR139. Increased miR-139 levels results in downregulation of EIF4G2, PTPRT, and HPGD and lead to cell death of MLL-AF9 cells.
Discussion
Our study has revealed key aspects of the molecular regulation of the tumor suppressor MIR139 in MLL-AF9 leukemia. We describe that expression of miR-139 in MLL-AF9 cells caused apoptosis, abolished colony formation in vitro, and inhibited MLL-AF9 leukemia in vivo. Besides the previously published target Eif4g2 [13], we identified Hpgd and Ptprt as novel miR-139-controlled targets of which depletion in MLL-AF9 AML at least in part phenocopied miR-139 expression. Our data suggest a mechanism of MIR139 silencing in MLL-AF9 AML in which PRC2 activity and recruitment of POLR2M at the E1, E2 inhibit MIR139 expression to promote oncogenesis (Fig. 7).
There is emerging evidence that MIR139 is a tumor suppressor in various types of solid cancer [15, 39] and leukemia [12,13,14]. Eif4g2 is a target of miR-139-5p in AML [13], which is confirmed in our study. The RNA-binding protein Elavl1 is commonly found as a target for miR-139-3p in solid cancer [39] and upon ICL-induced stress in HSPCs [12]. In the present study however, Elavl1 is not differentially expressed between HSPCs and MLL-AF9 cells and when miR-139 expression is induced in MLL-AF9-139 cells. This suggests that miR-139 dependent target regulation is cell type- and context-dependent, which has been previously observed for other miRNAs and cell types [40,41,42].
PRC2-mediated repression of tumor suppressor genes, including Cdkn2a (p16INK4A), Cdkn2b (p15INK4B), Gata2 and Egr1, is a critical event in MLL-AF9 AML [22, 23, 43]. We provide evidence that Mir139 is a tumor suppressor that is targeted by PRC2 downstream of MLL-AF9. First, MLL-AF9 expression in HSPCs caused strong repression of Mir139 via PRC2. This is a critical step in oncogenic transformation as we demonstrated that miR-139 induction in MLL-AF9 cells caused apoptosis, loss of colony-forming capacity and inhibition of leukemogenesis in mice. We found that escaping AML cells in the DOX-treated mice have a miR-139 upregulation of only less than eightfold due to unknown reasons, in contrast to the 50-100-fold upregulation as found in vitro experiments. Apparently, an eightfold induction of miR-139 is not sufficient to cause apoptosis in the transplanted AML cells, allowing this aberrant sub-clone to give rise to full-blown leukemia. Second, genetic Mir139 deletion partly rescued the oncolytic effect of PRC2 inhibition in MLL-AF9 cells. Notably, the enhanced expansion capacity of MLL-AF9-Mir139KO cells compared with MLL-AF9 cells indicate that complete loss of Mir139 is a selective advantage for leukemia outgrowth in vitro. These results explain clinical data showing that AML patients with the lowest miR-139 expression levels have a poor prognosis [13]. Third, inhibition of methyltransferases EZH1 and EZH2 by UNC1999 greatly induced miR-139 expression in MLL-AF9 AML patient samples and other subtypes of AML, demonstrating direct silencing of Mir139 by PRC2 in AML, which is in agreement with previous findings in solid cancers [44, 45]. This suggests that the PRC2-mediated MIR139 silencing is a general mechanism in cancer and may be a key event in oncogenesis. Fourth, we identified Eif4g2, Ptprt, and Hpgd as miR-139 targets, which are involved in MLL-AF9 cell survival. Deregulation of these three genes by mutation, disruption of miRNA-mediated regulation or genomic aberrations are frequently found in different types of cancer [13, 46,47,48,49,50]. Increased expression of Eif4g2, caused by MIR139 silencing, enhances protein synthesis [46], which is essential for the proliferation of AML cells [13]. Ptprt controls JAK/STAT signaling by dephosphorylation of STAT3, which is frequently dysregulated in human cancer, including AML [51, 52]. The expression and phosphorylation of STAT3 beta (STAT3β), which is an isoform of STAT3 has tumor-suppressive activities in MLL-AF9 AML [53]. Therefore, as a result of MIR139 silencing, STAT3β phosphorylation is potentially decreased, thereby contributing to MLL-AF9 oncogenesis. HPGD is an enzyme that converts PGE2 to 15-keto-PGE2 and has been associated with epithelial-to-mesenchymal transition and poor survival of patients with breast cancer and lung cancer [46, 54]. Furthermore, our proteomics data demonstrate HPGD as the most upregulated protein in MLL-AF9 cells compared with WT HSPCs, suggesting that PGE2 conversion by HPGD contributes to MLL-AF9 oncogenesis. At last, we further demonstrate that miR-139 expression in MLL-AF9-i139 cells induces loss of colony-forming capacity and apoptosis. Our gene expression data show that genes that inhibit cell cycle progression, such as Btg2 [55], Cdkn1a [56], and Cdkn2d [57], and genes that promote apoptosis such as Bmf [58], Dusp1 [59], Rnasel [60], Trp53inp1 [61], Ypel3 [62], and Zc3h12a [63] are upregulated upon miR-139 expression and may explain this phenotype. Taken together, we conclude that MIR139 is silenced via PRC2 and that this silencing mechanism induces different pathways via miR-139 targets that contribute to cell survival and MLL-AF9 leukemogenesis.
Our data indicate that Mir139 silencing is independent of the transcriptional regulation of Pde2a. This may be explained by the high percentage of methylated CpGs within E1 and E2 in MLL-AF9 cells, whereas the upstream promoter region is relatively less methylated. PRC2 prefers binding to methylated DNA via AEBP2 [34]. Accordingly, CpGs in the genomic regions overlapping with the POL-II binding sites in E1 and E2 were methylated in Molm-13 and THP-1 cells. In addition, we found that both enhancers are essential for PRC2-mediated Mir139 silencing in MLL-AF9 cells. Thus, CpG methylation at these enhancers may be involved in the selective repression of Mir139, leaving the Pde2a host gene expression intact.
We found that Pde2a expression and intron-1 splicing did not result in miR-139 expression in MLL-AF9 cells. These data suggest that in MLL-AF9 cells pri-miR-139 is degraded. We did not observe miR-139 induction upon PRC2 inhibition in MLL-AF9-E1KO cells and a reduced miR-139 induction in MLL-AF9-E2KO, suggesting that factors involved in miR-139 stability and processing may be recruited to the enhancers. Enhancer regions may play critical roles in the transcriptional regulation of miRNAs in cancer [64]. A recent study showed that DROSHA and DGCR8 interact with super-enhancer regions and boost miRNA production [65]. In agreement with these studies, our data suggest a mechanism in which the RNA-processing machinery is directly coupled to enhancer-mediated transcription of MIR139. This interaction may occur under cellular stress conditions resulting in increased miR-139 expression and cell death to prevent oncogenic transformation.
POL-II is the key factor responsible for the transcription of miRNAs [66]. Although stable paused transcription seems a logical downstream consequence of PcG-mediated gene silencing [67], the factors involved in the molecular mechanism of PRC2-mediated transcriptional pausing remained unknown. We found that POLR2M is recruited in a PRC2-dependent fashion to E1 and E2 to silence MIR139 expression. POLR2M interacts with the POL-II complex to pause transcription [38, 68, 69]. Promoter-proximal pausing of POL-II at TSSs has been correlated with H3K27me3 and PcG-silenced genes [70,71,72]. Paused POL-II complexes have also been found at enhancer regions [73]. Together, our study indicates that POLR2M interacts with E1 and E2 and silences MIR139 expression in MLL-AF9.
Our data demonstrate in different ways that POLR2M is a direct silencer of MIR139 in AML. First, POLR2M depletion in MLL-AF9 patient samples and human AML cell lines induced the expression of pri-miR-139. Although the levels of the mature miR-139-5p and miR-139-3p are low compared with experiments with UNC1999-mediated repression of PRC2, this could be explained by the fact that the CRISPR-Cas9-mediated depletion of POLR2M by lentiviral transduction is an asynchronous process. This causes a heterogeneous induction of MIR139 expression and subsequent cell death, thereby underestimating miR-139 levels. Second, POLR2M-depleted MLL-AF9 cells were lost in our CRISPR-Cas9 screen and validation experiments, whereas POLR2M-depleted MLL-AF9-Mir139KO cells survived, indicating that POLR2M-controlled survival is specifically mediated by the silencing of MIR139. Third, we showed that POLR2M is bound to the E1 and E2 regions and TSS of MIR139. Depletion of POLR2M at these loci was observed as a result of UNC1999-mediated inhibition of PRC2 and coincides with induction of miR-139 expression and cell death.
In conclusion, we have identified key molecular mechanisms that control tumor suppressor MIR139 in MLL-AF9 leukemia and report functionally relevant targets of miR-139-mediated cell death of MLL-AF9 cells. Furthermore, our findings reveal a PRC2-dependent POLR2M-mediated silencing mechanism of the MIR139 tumor suppressor in MLL-AF9 leukemia. The specific interaction of POLR2M with the POL-II complex is essential for transcriptional inhibition [74]. Therefore, POLR2M would be interesting for therapeutic targeting, for instance by molecularly interfering with the interaction with POL-II, thereby activating MIR139 expression and eliminating AML. Together, our findings highlight the importance of POLR2M-mediated silencing of MIR139 in MLL-AF9 AML.
Data availability
The NGS data generated in this paper is available from the GEO database with the following identifiers. GSE160404: RNA-seq of WT HSPCs, Mir139KO HSPCs and MLL-AF9 cells GSE160405: RNA-seq of MLL-AF9-i139 cells treated with and without DOX GSE160403: DNA-seq of MLL-AF9 WT and MLL-AF9 Mir139KO cells transduced with LentiCRISPRv2 GeCKOv2 sgRNA library. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD022488.
References
Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301.
Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114:2489–96.
Winters AC, Bernt KM. MLL-rearranged leukemias-an update on science and clinical approaches. Front Pediatr. 2017;5:4.
Steinhilber D, Marschalek R. How to effectively treat acute leukemia patients bearing MLL-rearrangements? Biochem Pharm. 2018;147:183–90.
Kotani S, Yoda A, Kon A, Kataoka K, Ochi Y, Shiozawa Y, et al. Molecular pathogenesis of disease progression in MLL-rearranged AML. Leukemia. 2019;33:612–24.
Xu J, Li L, Xiong J, denDekker A, Ye A, Karatas H, et al. MLL1 and MLL1 fusion proteins have distinct functions in regulating leukemic transcription program. Cell Discov. 2016;2:16008.
Stavropoulou V, Kaspar S, Brault L, Sanders MA, Juge S, Morettini S, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive aml expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30:43–58.
Horton SJ, Jaques J, Woolthuis C, van Dijk J, Mesuraca M, Huls G, et al. MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia. 2013;27:1116–26.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838.
Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–85.
Alemdehy MF, Haanstra JR, de Looper HW, van Strien PM, Verhagen-Oldenampsen J, Caljouw Y, et al. ICL-induced miR139-3p and miR199a-3p have opposite roles in hematopoietic cell expansion and leukemic transformation. Blood. 2015;125:3937–48.
Emmrich S, Engeland F, El-Khatib M, Henke K, Obulkasim A, Schoning J, et al. miR-139-5p controls translation in myeloid leukemia through EIF4G2. Oncogene. 2016;35:1822–31.
Krowiorz K, Ruschmann J, Lai C, Ngom M, Maetzig T, Martins V, et al. MiR-139-5p is a potent tumor suppressor in adult acute myeloid leukemia. Blood Cancer J. 2016;6:e508.
Bao W, Fu HJ, Xie QS, Wang L, Zhang R, Guo ZY, et al. HER2 interacts with CD44 to up-regulate CXCR4 via epigenetic silencing of microRNA-139 in gastric cancer cells. Gastroenterology. 2011;141:2076–2087 e2076.
Wang W, Qin JJ, Voruganti S, Nag S, Zhou J, Zhang R. Polycomb group (PcG) proteins and human cancers: multifaceted functions and therapeutic implications. Med Res Rev. 2015;35:1220–67.
Huang X, Yan J, Zhang M, Wang Y, Chen Y, Fu X, et al. Targeting epigenetic crosstalk as a therapeutic strategy for EZH2-aberrant solid tumors. Cell. 2018;175:186–199 e119.
Zhang H, Zhu D, Zhang Z, Kaluz S, Yu B, Devi NS, et al. EZH2 targeting reduces medulloblastoma growth through epigenetic reactivation of the BAI1/p53 tumor suppressor pathway. Oncogene. 2020;39:1041–1048.
van Mierlo G, Veenstra GJC, Vermeulen M, Marks H. The complexity of PRC2 subcomplexes. Trends Cell Biol. 2019;29(Aug):660–71.
Laugesen A, Hojfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell. 2019;74(Apr 4):8–18.
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002;16:2893–905.
Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci USA. 2012;109:5028–5033.
Danis E, Yamauchi T, Echanique K, Haladyna J, Kalkur R, Riedel S, et al. Inactivation of Eed impedes MLL-AF9-mediated leukemogenesis through Cdkn2a-dependent and Cdkn2a-independent mechanisms in a murine model. Exp Hematol. 2015;43:930–935 e936.
Aranda S, Mas G, Di, Croce L. Regulation of gene transcription by polycomb proteins. Sci Adv. 2015;1:e1500737.
Hope JH, Zhao M, Stairiker CJ, Kiernan C, Alison CJ, Mueller YM, et al. MicroRNA-139 expression is dispensable for the generation of influenza-specific CD8+ T cell responses. J Immunol 2021 In Press.
van Dongen JJ, Lhermitte L, Bottcher S, Almeida J, van der Velden VH, Flores-Montero J, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26:1908–1975.
Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922.
Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828.
Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125:346–357.
Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015;27:589–602.
Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949.
Volk A, Liang K, Suraneni P, Li X, Zhao J, Bulic M, et al. A CHAF1B-dependent molecular switch in hematopoiesis and leukemia pathogenesis. Cancer Cell. 2018;34:707–723 e707.
Ordonez R, Martinez-Calle N, Agirre X, Prosper F. DNA methylation of enhancer elements in myeloid neoplasms: think outside the promoters? Cancers (Basel) 2019;11:1424.
Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW, et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017;24:1028–1038.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784.
DeLaney E, Luse DS. Gdown1 associates efficiently with RNA polymerase II after promoter clearance and displaces TFIIF during transcript elongation. PLoS One. 2016;11:e0163649.
Guo J, Turek ME, Price DH. Regulation of RNA polymerase II termination by phosphorylation of Gdown1. J Biol Chem. 2014;289:12657–12665.
Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, et al. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol Cell. 2012;45:51–63.
Ni ZZ, He JK, Tang X, Tao Z, Zhang Y, Xie B. Identification of ELAVL1 gene and miRNA-139-3p involved in the aggressiveness of NSCLC. Eur Rev Med Pharm Sci. 2020;24:9453–9464.
Kohram F, Fallah P, Shamsara M, Bolandi Z, Rassoulzadegan M, Soleimani M, et al. Cell type-dependent functions of microRNA-92a. J Cell Biochem. 2018;119:5798–5804.
Hsin JP, Lu Y, Loeb GB, Leslie CS, Rudensky AY. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat Immunol. 2018;19:1137–1145.
Nussbacher JK, Yeo GW. Systematic discovery of RNA binding proteins that regulate MicroRNA levels. Mol Cell. 2018;69:1005–1016 e1007.
Shi J, Wang E, Zuber J, Rappaport A, Taylor M, Johns C, et al. The polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2013;32:930–938.
Ma J, Zhang J, Weng YC, Wang JC. EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal transition and lymph node metastasis of pancreatic cancer. Mol Cells. 2018;41:868–880.
Wu D, Wu F, Li B, Huang W, Wang D. EZH2 promotes the expression of LPA1 by mediating microRNA-139 promoter methylation to accelerate the development of ovarian cancer. Cancer Cell Int. 2020;20:551.
Lehtinen L, Vainio P, Wikman H, Reemts J, Hilvo M, Issa R, et al. 15-Hydroxyprostaglandin dehydrogenase associates with poor prognosis in breast cancer, induces epithelial-mesenchymal transition, and promotes cell migration in cultured breast cancer cells. J Pathol. 2012;226:674–686.
He N, Zheng H, Li P, Zhao Y, Zhang W, Song F, et al. miR-485-5p binding site SNP rs8752 in HPGD gene is associated with breast cancer risk. PLoS One. 2014;9:e102093.
Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166.
Ku M, MacKinnon RN, Wall M, Narayan N, Walkley C, Cheng HC, et al. Hemopoietic Cell Kinase amplification with Protein Tyrosine Phosphatase Receptor T depletion leads to polycythemia, aberrant marrow erythoid maturation, and splenomegaly. Sci Rep. 2019;9:7050.
Yao Y, Shao J, Wu J, Zhang Q, Wang J, Xiao D, et al. The functional variant in the 3’UTR of PTPRT with the risk of esophageal squamous cell carcinoma in a chinese population. Cell Physiol Biochem. 2015;36:306–314.
Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood. 2011;117:5701–5709.
Sen M, Kindsfather A, Danilova L, Zhang F, Colombo R, LaPorte MG, et al. PTPRT epigenetic silencing defines lung cancer with STAT3 activation and can direct STAT3 targeted therapies. Epigenetics. 2020;15:604–617.
Aigner P, Mizutani T, Horvath J, Eder T, Heber S, Lind K, et al. STAT3beta is a tumor suppressor in acute myeloid leukemia. Blood Adv. 2019;3:1989–2002.
Tai HH, Tong M, Ding Y. 15-hydroxyprostaglandin dehydrogenase (15-PGDH) and lung cancer. Prostaglandins Other Lipid Mediat. 2007;83:203–208.
Yuniati L, Scheijen B, van der Meer LT, van Leeuwen FN. Tumor suppressors BTG1 and BTG2: beyond growth control. J Cell Physiol. 2019;234:5379–5389.
Kleinsimon S, Longmuss E, Rolff J, Jager S, Eggert A, Delebinski C, et al. GADD45A and CDKN1A are involved in apoptosis and cell cycle modulatory effects of viscumTT with further inactivation of the STAT3 pathway. Sci Rep. 2018;8:5750.
Wang Y, Jin W, Jia X, Luo R, Tan Y, Zhu X, et al. Transcriptional repression of CDKN2D by PML/RARalpha contributes to the altered proliferation and differentiation block of acute promyelocytic leukemia cells. Cell Death Dis. 2014;5:e1431.
Grespi F, Soratroi C, Krumschnabel G, Sohm B, Ploner C, Geley S, et al. BH3-only protein Bmf mediates apoptosis upon inhibition of CAP-dependent protein synthesis. Cell Death Differ. 2010;17:1672–1683.
Yang J, Sun L, Han J, Zheng W, Peng W. DUSP1/MKP-1 regulates proliferation and apoptosis in keratinocytes through the ERK/Elk-1/Egr-1 signaling pathway. Life Sci. 2019;223:47–53.
Diaz-Guerra M, Rivas C, Esteban M. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology. 1997;236:354–363.
Ishii K, Ishiai M, Morimoto H, Kanatsu-Shinohara M, Niwa O, Takata M, et al. The Trp53-Trp53inp1-Tnfrsf10b pathway regulates the radiation response of mouse spermatogonial stem cells. Stem Cell Rep. 2014;3:676–689.
Baker SJ. Small unstable apoptotic protein, an apoptosis-associated protein, suppresses proliferation of myeloid cells. Cancer Res. 2003;63:705–712.
Mao R, Yang R, Chen X, Harhaj EW, Wang X, Fan Y. Regnase-1, a rapid response ribonuclease regulating inflammation and stress responses. Cell Mol Immunol. 2017;14:412–422.
Tang F, Zhang Y, Huang QQ, Qian MM, Li ZX, Li YJ, et al. Genome-wide identification and analysis of enhancer-regulated microRNAs across 31 human cancers. Front Genet. 2020;11:644.
Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated RNA processing revealed by integrative MicroRNA network analysis. Cell. 2017;168:1000–1014 e1015.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297.
Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577.
Jishage M, Ito K, Chu CS, Wang X, Yamaji M, Roeder RG. Transcriptional down-regulation of metabolic genes by Gdown1 ablation induces quiescent cell re-entry into the cell cycle. Genes Dev. 2020;34:767–784.
Jishage M, Yu X, Shi Y, Ganesan SJ, Chen WY, Sali A, et al. Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1. Nat Struct Mol Biol. 2018;25:859–867.
Liu J, Wu X, Zhang H, Pfeifer GP, Lu Q. Dynamics of RNA polymerase II pausing and bivalent histone h3 methylation during neuronal differentiation in brain development. Cell Rep. 2017;20:1307–1318.
Mantsoki A, Devailly G, Joshi A. Dynamics of promoter bivalency and RNAP II pausing in mouse stem and differentiated cells. BMC Dev Biol. 2018;18:2.
Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754.
Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018;32:26–41.
Jishage M, Roeder RG. Regulation of hepatocyte cell cycle re-entry by RNA polymerase II-associated Gdown1. Cell Cycle. 2020;19:3222–3230.
Acknowledgements
For this work, C.J.S. and I.V.Z. were supported by the Dutch Cancer Foundation (KWF), grant no.: 10948. Furthermore, the Dutch Research Council (NWO) Medium Investment Grant 91116004 (partly financed by ZonMw) to P.A.v.V. also supported the work. Furthermore, we would like to thank Yvette Caljouw, Fabiënne van Opstal, Bernard Stikker and Stijn van den Broek for their technical assistance and Dr. G. Dal Collo and Dr. C. Kiernan for critically reviewing the manuscript. In addition, we would like to acknowledge Dr. R. Delwel and Dr. R. Avelino for their experimental and technical advice.
Author information
Authors and Affiliations
Contributions
C.J.S., P.D.K., and S.J.E. wrote the manuscript. S.J.E. designed the project. C.J.S., I.V.Z., A.Ö., and A.V.H.B. performed most of the experiments. G.M.C.J. and P.A.V.V. performed the proteomic analysis. E.B. performed next-generation sequencing (NGS). L.G.L. performed the bioinformatics analysis of the RNA-sequence data. C.J.S., C.S., and J.S.A.V. performed CRISPR-KO screens. R.W.W.B. and W.F.J. performed CRISPR library sequencing and bioinformatic analysis. A.M. generated the E1KO and E2KO mouse models. V.H.J.vd V. wrote the METC protocol and collected human MLL-AF9 patient samples. E.K., M.B., and M.A.J. performed the mouse leukemia and MLL-AF9 stem cell experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stavast, C.J., van Zuijen, I., Karkoulia, E. et al. The tumor suppressor MIR139 is silenced by POLR2M to promote AML oncogenesis. Leukemia 36, 687–700 (2022). https://doi.org/10.1038/s41375-021-01461-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01461-5