Abstract
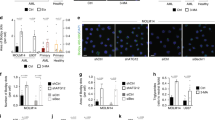
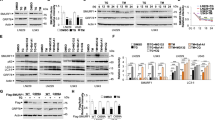
Sphingolipids and their metabolic pathways have been implicated in disease development and therapeutic response; however, the detailed mechanisms remain unclear. Using a sphingolipid network focused CRISPR/Cas9 library screen, we identified an endoplasmic reticulum (ER) enzyme, 3-Ketodihydrosphingosine reductase (KDSR), to be essential for leukemia cell maintenance. Loss of KDSR led to apoptosis, cell cycle arrest, and aberrant ER structure. Transcriptomic analysis revealed the indispensable role of KDSR in maintaining the unfolded protein response (UPR) in ER. High-density CRISPR tiling scan and sphingolipid mass spectrometry pinpointed the critical role of KDSR’s catalytic function in leukemia. Mechanistically, depletion of KDSR resulted in accumulated 3-ketodihydrosphingosine (KDS) and dysregulated UPR checkpoint proteins PERK, ATF6, and ATF4. Finally, our study revealed the synergism between KDSR suppression and pharmacologically induced ER-stress, underscoring a therapeutic potential of combinatorial targeting sphingolipid metabolism and ER homeostasis in leukemia treatment.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Code availability
The computational codes/tool packages used in this study are available through developers and venders, including Genetic Perturbation Platform, CLC Main Workbench, Bowtie2, and Gaussian kernel smoothing in R.
References
Warner JK, Wang JC, Hope KJ, Jin L, Dick JE. Concepts of human leukemic development. Oncogene. 2004;23:7164–77.
Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501.
Yu J, Jiang PYZ, Sun H, Zhang X, Jiang Z, Li Y, et al. Advances in targeted therapy for acute myeloid leukemia. Biomark Res. 2020;8:17.
Zjablovskaja P, Florian MC Acute Myeloid Leukemia: Aging and Epigenetics. Cancers (Basel). 2019;12:103.
Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18:33–50.
Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science 2010;327:46–50.
Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:175–91.
Lewis AC, Wallington-Beddoe CT, Powell JA, Pitson SM. Targeting sphingolipid metabolism as an approach for combination therapies in haematological malignancies. Cell Death Disco. 2018;4:4.
Bezombes C, de Thonel A, Apostolou A, Louat T, Jaffrezou JP, Laurent G, et al. Overexpression of protein kinase Czeta confers protection against antileukemic drugs by inhibiting the redox-dependent sphingomyelinase activation. Mol Pharm. 2002;62:1446–55.
El-Assaad W, Kozhaya L, Araysi S, Panjarian S, Bitar FF, Baz E, et al. Ceramide and glutathione define two independently regulated pathways of cell death initiated by p53 in Molt-4 leukaemia cells. Biochem J. 2003;376:725–32.
Jarvis WD, Fornari FA Jr., Browning JL, Gewirtz DA, Kolesnick RN, Grant S. Attenuation of ceramide-induced apoptosis by diglyceride in human myeloid leukemia cells. J Biol Chem. 1994;269:31685–92.
Wang H, Maurer BJ, Liu YY, Wang E, Allegood JC, Kelly S, et al. N-(4-Hydroxyphenyl)retinamide increases dihydroceramide and synergizes with dimethylsphingosine to enhance cancer cell killing. Mol Cancer Ther. 2008;7:2967–76.
Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, et al. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. J Biol Chem. 2007;282:10922–34.
Tan SF, Liu X, Fox TE, Barth BM, Sharma A, Turner SD, et al. Acid ceramidase is upregulated in AML and represents a novel therapeutic target. Oncotarget. 2016;7:83208–22.
Wallington-Beddoe CT, Powell JA, Tong D, Pitson SM, Bradstock KF, Bendall LJ. Sphingosine kinase 2 promotes acute lymphoblastic leukemia by enhancing MYC expression. Cancer Res. 2014;74:2803–15.
Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci. 2016;73:79–94.
Bhattarai KR, Chaudhary M, Kim HR, Chae HJ. Endoplasmic Reticulum (ER) Stress Response Failure in Diseases. Trends Cell Biol. 2020;30:672–5.
Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29.
Tanimura A, Yujiri T, Tanaka Y, Hatanaka M, Mitani N, Nakamura Y, et al. The anti-apoptotic role of the unfolded protein response in Bcr-Abl-positive leukemia cells. Leuk Res. 2009;33:924–8.
Sudsaward S, Khunchai S, Thepmalee C, Othman A, Limjindaporn T, Yenchitsomanus PT, et al. Endoplasmic reticulum stress, unfolded protein response and autophagy contribute to resistance to glucocorticoid treatment in human acute lymphoblastic leukaemia cells. Int J Oncol. 2020;57:835–44.
Zhou C, Martinez E, Di Marcantonio D, Solanki-Patel N, Aghayev T, Peri S, et al. JUN is a key transcriptional regulator of the unfolded protein response in acute myeloid leukemia. Leukemia. 2017;31:1196–205.
Martelli AM, Paganelli F, Chiarini F, Evangelisti C, McCubrey JA. The unfolded protein response: a novel therapeutic target in acute leukemias. Cancers (Basel). 2020;12:333.
Masciarelli S, Capuano E, Ottone T, Divona M, Lavorgna S, Liccardo F, et al. Retinoic acid synergizes with the unfolded protein response and oxidative stress to induce cell death in FLT3-ITD+ AML. Blood Adv. 2019;3:4155–60.
Goda AE, Erikson RL, Sakai T, Ahn JS, Kim BY. Preclinical evaluation of bortezomib/dipyridamole novel combination as a potential therapeutic modality for hematologic malignancies. Mol Oncol. 2015;9:309–22.
Kihara A, Igarashi Y. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J Biol Chem. 2004;279:49243–50.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 2015;527:192–7.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Mascorro JA, Bozzola JJ. Processing biological tissues for ultrastructural study. Methods Mol Biol. 2007;369:19–34.
Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524–8.
Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 2016;167:1867–82 e21.
Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89.
Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, et al. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Disco. 2016;6:900–13.
Schoonenberg VAC, Cole MA, Yao Q, Macias-Trevino C, Sher F, Schupp PG, et al. CRISPRO: identification of functional protein coding sequences based on genome editing dense mutagenesis. Genome Biol. 2018;19:169.
Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–7.
Rosati E, Sabatini R, Rampino G, De Falco F, Di Ianni M, Falzetti F, et al. Novel targets for endoplasmic reticulum stress-induced apoptosis in B-CLL. Blood 2010;116:2713–23.
Morin MJ, Porter CW, McKernan P, Bernacki RJ. The biochemical and ultrastructural effects of tunicamycin and D-glucosamine in L1210 leukemic cells. J Cell Physiol. 1983;114:162–72.
Lehle L, Tanner W. The specific site of tunicamycin inhibition in the formation of dolichol-bound N-acetylglucosamine derivatives. FEBS Lett. 1976;72:167–70.
Yoo J, Mashalidis EH, Kuk ACY, Yamamoto K, Kaeser B, Ichikawa S, et al. GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation. Nat Struct Mol Biol. 2018;25:217–24.
Chan AKN, Chen CW. Rewiring the Epigenetic Networks in MLL-Rearranged Leukemias: Epigenetic Dysregulation and Pharmacological Interventions. Front Cell Dev Biol. 2019;7:81.
Federici L, Falini B. Nucleophosmin mutations in acute myeloid leukemia: a tale of protein unfolding and mislocalization. Protein Sci. 2013;22:545–56.
Schardt JA, Mueller BU, Pabst T. Activation of the unfolded protein response in human acute myeloid leukemia. Methods Enzymol. 2011;489:227–43.
Haefliger S, Klebig C, Schaubitzer K, Schardt J, Timchenko N, Mueller BU, et al. Protein disulfide isomerase blocks CEBPA translation and is up-regulated during the unfolded protein response in AML. Blood 2011;117:5931–40.
Giuli MV, Diluvio G, Giuliani E, Franciosa G, Di Magno L, Pignataro MG, et al. Notch3 contributes to T-cell leukemia growth via regulation of the unfolded protein response. Oncogenesis 2020;9:93.
Kharabi Masouleh B, Geng H, Hurtz C, Chan LN, Logan AC, Chang MS, et al. Mechanistic rationale for targeting the unfolded protein response in pre-B acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2014;111:E2219–28.
Sun H, Lin DC, Guo X, Kharabi Masouleh B, Gery S, Cao Q, et al. Inhibition of IRE1alpha-driven pro-survival pathways is a promising therapeutic application in acute myeloid leukemia. Oncotarget 2016;7:18736–49.
Pellegrini P, Selvaraju K, Faustini E, Mofers A, Zhang X, Ternerot J, et al. Induction of ER stress in acute lymphoblastic leukemia cells by the deubiquitinase inhibitor VLX1570. Int J Mol Sci. 2020;21:4757.
Kharabi Masouleh B, Chevet E, Panse J, Jost E, O’Dwyer M, Bruemmendorf TH, et al. Drugging the unfolded protein response in acute leukemias. J Hematol Oncol. 2015;8:87.
Zi J, Han Q, Gu S, McGrath M, Kane S, Song C, et al. Targeting NAT10 induces apoptosis associated with enhancing endoplasmic reticulum stress in acute myeloid leukemia cells. Front Oncol. 2020;10:598107.
Krebs S, Medugorac I, Rother S, Strasser K, Forster M. A missense mutation in the 3-ketodihydrosphingosine reductase FVT1 as candidate causal mutation for bovine spinal muscular atrophy. Proc Natl Acad Sci USA. 2007;104:6746–51.
Takeichi T, Torrelo A, Lee JYW, Ohno Y, Lozano ML, Kihara A, et al. Biallelic mutations in KDSR disrupt ceramide synthesis and result in a spectrum of keratinization disorders associated with thrombocytopenia. J Invest Dermatol. 2017;137:2344–53.
Bariana TK, Labarque V, Heremans J, Thys C, De Reys M, Greene D, et al. Sphingolipid dysregulation due to lack of functional KDSR impairs proplatelet formation causing thrombocytopenia. Haematologica 2019;104:1036–45.
Boyden LM, Vincent NG, Zhou J, Hu R, Craiglow BG, Bayliss SJ, et al. Mutations in KDSR cause recessive progressive symmetric erythrokeratoderma. Am J Hum Genet. 2017;100:978–84.
Huber M, Chiticariu E, Bachmann D, Flatz L, Hohl D. Palmoplantar keratoderma with leukokeratosis anogenitalis caused by KDSR mutations. J Invest Dermatol. 2020;140:1662–5 e1.
Park KH, Ye ZW, Zhang J, Hammad SM, Townsend DM, Rockey DC, et al. 3-ketodihydrosphingosine reductase mutation induces steatosis and hepatic injury in zebrafish. Sci Rep. 2019;9:1138.
Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–40.
Akiyama M, Hatanaka M, Ohta Y, Ueda K, Yanai A, Uehara Y, et al. Increased insulin demand promotes while pioglitazone prevents pancreatic beta cell apoptosis in Wfs1 knockout mice. Diabetologia 2009;52:653–63.
Esk C, Lindenhofer D, Haendeler S, Wester RA, Pflug F, Schroeder B, et al. A human tissue screen identifies a regulator of ER secretion as a brain-size determinant. Science 2020;370:935–41.
Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 2009;460:978–83.
Moss TJ, Andreazza C, Verma A, Daga A, McNew JA. Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc Natl Acad Sci USA. 2011;108:11133–8.
Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102.
Tam AB, Roberts LS, Chandra V, Rivera IG, Nomura DK, Forbes DJ, et al. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev Cell. 2018;46:327–43 e7.
Acknowledgements
This work was supported by the American Society of Hematology (ASH Scholar Award to C.-W.C.), Alex’s Lemonade Stand Foundation (ALSF Innovation Award to C.-W.C.), National Institutes of Health Grants CA197498, CA233691, CA236626 (to C.-W.C.), Graduate Academic Exchange Scholarship of Fujian Medical University (to Q.L.), and Riggs-Union International Exchange Scholarship of Fujian Medical University Union Hospital (to Q.L.). Research reported in this publication included work performed in the City of Hope’s Mass Spectrometry and Proteomics Core Facility (Dr. Gabriel Gugiu) and Electron Microscopy Core Facility (Drs. Zhuo Li and Ricardo Zerda) supported by the National Cancer Institute of the National Institutes of Health P30 Grant CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
QL, AKNC, W-HC, LY, SPP, KM, NM, XX, ML performed the experiments; QL, LY, AKNC, WL and C-WC analyzed the data; RJL, SYW, and C-WC provided conceptual input; QL, RJL, and C-WC wrote the paper; SYW and C-WC conceived and supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, Q., Chan, A.K.N., Chang, WH. et al. 3-Ketodihydrosphingosine reductase maintains ER homeostasis and unfolded protein response in leukemia. Leukemia 36, 100–110 (2022). https://doi.org/10.1038/s41375-021-01378-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01378-z
This article is cited by
-
The molecular mechanisms underlying the regeneration process in the earthworm, Perionyx excavatus exhibit indications of apoptosis-induced compensatory proliferation (AICP)
In Vitro Cellular & Developmental Biology - Animal (2024)
-
A novel class of inhibitors that disrupts the stability of integrin heterodimers identified by CRISPR-tiling-instructed genetic screens
Nature Structural & Molecular Biology (2024)