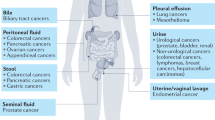
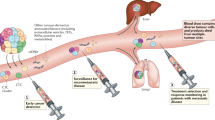
Abstract
Liquid biopsies—a source of circulating cell-free nucleic acids, proteins and extracellular vesicles—are currently being explored for the quantitative and qualitative characterisation of the tumour genome and as a mode of non-invasive therapeutic monitoring in cancer. Emerging data suggest that liquid biopsies might offer a potentially simple, non-invasive, repeatable strategy for diagnosis, prognostication and therapeutic decision making in a genetically heterogeneous disease like multiple myeloma (MM), with particular applicability in subsets of patients where conventional markers of disease burden may be less informative. In this review, we describe the emerging utility of the evaluation of circulating tumour DNA, extracellular RNA, cell-free proteins and metabolites and extracellular vesicles in MM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Change history
20 September 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41375-021-01399-8
References
Kumar SK, Dispenzieri A, Lacy MQ, Gertz MA, Buadi FK, Pandey S, et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28:1122–8.
Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101.
Mithraprabhu S, Khong T, Ramachandran M, Chow A, Klarica D, Mai L, et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia. 2017;31:1695–705.
Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun. 2017;8:268.
Weinhold N, Chavan SS, Rasche L, Stephens OW, Patel P, Tytarenko R, et al. Extensive regional intra-clonal heterogeneity in multiple myeloma—implications for diagnostics, risk stratification and targeted treatment. Blood. 2016;128:3278.
Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28:384–90.
Mithraprabhu S, Sirdesai S, Chen M, Khong T, Spencer A. Circulating tumour DNA analysis for tumour genome characterisation and monitoring disease burden in extramedullary multiple myeloma. Int J Mol Sci. 2018;19:1858.
Mithraprabhu S, Morley R, Khong T, Kalff A, Bergin K, Hocking J, et al. Monitoring tumour burden and therapeutic response through analysis of circulating tumour DNA and extracellular RNA in multiple myeloma patients. Leukemia. 2019;33:2022–33.
Chen M, Mithraprabhu S, Ramachandran M, Choi K, Khong T, Spencer A. Utility of circulating cell-free RNA analysis for the characterization of global transcriptome profiles of multiple myeloma patients. Cancers. 2019;11:887.
Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil. 1948;142:241–3.
Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra168.
Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12.
Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154.
Heitzer E, Auer M, Hoffmann EM, Pichler M, Gasch C, Ulz P, et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer. 2013;133:346–56.
Biancon G, Gimondi S, Vendramin A, Carniti C, Corradini P. Noninvasive molecular monitoring in multiple myeloma patients using cell-free tumor DNA: a pilot study. J Mol Diagn. 2018;20:859–70.
Manier S, Park J, Capelletti M, Bustoros M, Freeman SS, Ha G, et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat Commun. 2018;9:1691.
Guo G, Raje NS, Seifer C, Kloeber J, Isenhart R, Ha G, et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia. 2018;32:1838–41.
Deshpande S, Tytarenko RG, Wang Y, Boyle EM, Ashby C, Schinke CD, et al. Monitoring treatment response and disease progression in myeloma with circulating cell-free DNA. Eur J Haematol. 2021;106:230–40.
Manzoni M, Pompa A, Fabris S, Pelizzoni F, Ciceri G, Seia M, et al. Limits and applications of genomic analysis of circulating tumor DNA as a liquid biopsy in asymptomatic forms of multiple myeloma. Hemasphere. 2020;4:e402.
Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30.
Xu C, Nezami Ranjbar MR, Wu Z, DiCarlo J, Wang Y. Detecting very low allele fraction variants using targeted DNA sequencing and a novel molecular barcode-aware variant caller. BMC Genom. 2017;18:5.
Xu C, Gu X, Padmanabhan R, Wu Z, Peng Q, DiCarlo J, et al. smCounter2: an accurate low-frequency variant caller for targeted sequencing data with unique molecular identifiers. Bioinformatics. 2019;35:1299–309.
Mithraprabhu S, Hocking J, Ramachandran M, Choi K, Klarica D, Khong T, et al. DNA-repair gene mutations are highly prevalent in circulating tumour DNA from multiple myeloma patients. Cancers. 2019;11:917.
Kis O, Kaedbey R, Chow S, Danesh A, Dowar M, Li T, et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun. 2017;8:15086.
Oberle A, Brandt A, Voigtlaender M, Thiele B, Radloff J, Schulenkorf A, et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica. 2017;102:1105–11.
Mazzotti C, Buisson L, Maheo S, Perrot A, Chretien ML, Leleu X, et al. Myeloma MRD by deep sequencing from circulating tumor DNA does not correlate with results obtained in the bone marrow. Blood Adv. 2018;2:2811–3.
Rustad EH, Coward E, Skytoen ER, Misund K, Holien T, Standal T, et al. Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica. 2017;102:1266–72.
Li Q, Huang HJ, Ma J, Wang Y, Cao Z, Karlin-Neumann G, et al. RAS/RAF mutations in tumor samples and cell-free DNA from plasma and bone marrow aspirates in multiple myeloma patients. J Cancer. 2020;11:3543–50.
Long X, Xu Q, Lou Y, Li C, Gu J, Cai H, et al. The utility of non-invasive liquid biopsy for mutational analysis and minimal residual disease assessment in extramedullary multiple myeloma. Br J Haematol. 2020;189:e45–e48.
Vrabel D, Sedlarikova L, Besse L, Rihova L, Bezdekova R, Almasi M, et al. Dynamics of tumor-specific cfDNA in response to therapy in multiple myeloma patients. Eur J Haematol. 2020;104:190–7.
Mithraprabhu S, Spencer A. Circulating tumour DNA analysis in multiple myeloma. Oncotarget. 2017;8:90610–1.
Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162.
Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10:eaat4921.
Rawstron AC, Child JA, de Tute RM, Davies FE, Gregory WM, Bell SE, et al. Minimal residual disease assessed by multiparameter flow cytometry in multiple myeloma: impact on outcome in the Medical Research Council Myeloma IX Study. J Clin Oncol. 2013;31:2540–7.
Rawstron AC, Gregory WM, de Tute RM, Davies FE, Bell SE, Drayson MT, et al. Minimal residual disease in myeloma by flow cytometry: independent prediction of survival benefit per log reduction. Blood. 2015;125:1932–5.
Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil. 1948;142:241–3.
Mithraprabhu S, Spencer A. Liquid biopsy in multiple myeloma. https://doi.org/10.5772/intechopen.72652. IntechOpen, 2017.
Hao M, Zang M, Wendlandt E, Xu Y, An G, Gong D, et al. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int J Cancer. 2015;136:1835–44.
Qu X, Zhao M, Wu S, Yu W, Xu J, Xu J, et al. Circulating microRNA 483-5p as a novel biomarker for diagnosis survival prediction in multiple myeloma. Med Oncol. 2014;31:219.
Jones CI, Zabolotskaya MV, King AJ, Stewart HJ, Horne GA, Chevassut TJ, et al. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J Cancer. 2012;107:1987–96.
Huang JJ, Yu J, Li JY, Liu YT, Zhong RQ. Circulating microRNA expression is associated with genetic subtype and survival of multiple myeloma. Med Oncol. 2012;29:2402–8.
Navarro A, Diaz T, Tovar N, Pedrosa F, Tejero R, Cibeira MT, et al. A serum microRNA signature associated with complete remission and progression after autologous stem-cell transplantation in patients with multiple myeloma. Oncotarget. 2015;6:1874–83.
Besse L, Sedlarikova L, Kryukov F, Nekvindova J, Radova L, Slaby O, et al. Circulating serum MicroRNA-130a as a novel putative marker of extramedullary myeloma. PLoS One. 2015;10:e0137294.
Yoshizawa S, Ohyashiki JH, Ohyashiki M, Umezu T, Suzuki K, Inagaki A, et al. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J. 2012;2:e53.
Kubiczkova L, Kryukov F, Slaby O, Dementyeva E, Jarkovsky J, Nekvindova J, et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2014;99:511–8.
Jung SH, Lee SE, Lee M, Kim SH, Yim SH, Kim TW, et al. Circulating microRNA expressions can predict the outcome of lenalidomide plus low-dose dexamethasone treatment in patients with refractory/relapsed multiple myeloma. Haematologica. 2017;102:e459.
Gupta N, Kumar R, Seth T, Garg B, Sati HC, Sharma A. Clinical significance of circulatory microRNA-203 in serum as novel potential diagnostic marker for multiple myeloma. J Cancer Res Clin Oncol. 2019;145:1601–11.
Zhong L, Jin X, Xu Z, Zeng M, Chen D, He Y, et al. Circulating miR-451a levels as a potential biomarker to predict the prognosis of patients with multiple myeloma. Oncol Lett. 2020;20:263.
Rocci A, Hofmeister CC, Geyer S, Stiff A, Gambella M, Cascione L, et al. Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia. 2014;28:1922–6.
Hao M, Zang M, Zhao L, Deng S, Xu Y, Qi F, et al. Serum high expression of miR-214 and miR-135b as novel predictor for myeloma bone disease development and prognosis. Oncotarget. 2016;7:19589–600.
Zhu B, Chen H, Zhang X, Pan Y, Jing R, Shen L, et al. Serum miR-30d as a novel biomarker for multiple myeloma and its antitumor role in U266 cells through the targeting of the MTDH/PI3K/Akt signaling pathway. Int J Oncol. 2018;53:2131–44.
Fiehn O. Metabolomics-the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–71.
Ho M, Bianchi G, Anderson KC. Proteomics-inspired precision medicine for treating and understanding multiple myeloma. Expert Rev Precis Me. 2020;5:67–85.
Peng B, Li H, Peng XX. Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein Cell. 2015;6:628–37.
Marshall DD, Powers R. Beyond the paradigm: combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog Nucl Magn Reson Spectrosc. 2017;100:1–16.
Sobsey CA, Ibrahim S, Richard VR, Gaspar V, Mitsa G, Lacasse V, et al. Targeted and untargeted proteomics approaches in biomarker development. Proteomics. 2020;20:e1900029.
Janker L, Mayer RL, Bileck A, Kreutz D, Mader JC, Utpatel K, et al. Metabolic, anti-apoptotic and immune evasion strategies of primary human myeloma cells indicate adaptations to hypoxia. Mol Cell Proteom. 2019;18:936–53.
Ludwig C, Williams DS, Bartlett DB, Essex SJ, McNee G, Allwood JW, et al. Alterations in bone marrow metabolism are an early and consistent feature during the development of MGUS and multiple myeloma. Blood. Cancer J. 2015;5:e359.
Masarwi M, DeSchiffart A, Ham J, Reagan MR. Multiple myeloma and fatty acid metabolism. JBMR. 2019;3:e10173.
Puchades-Carrasco L, Lecumberri R, Martinez-Lopez J, Lahuerta JJ, Mateos MV, Prosper F, et al. Multiple myeloma patients have a specific serum metabolomic profile that changes after achieving complete remission. Clin Cancer Res. 2013;19:4770–9.
Deulofeu M, Kolarova L, Salvado V, Maria Pena-Mendez E, Almasi M, Stork M, et al. Rapid discrimination of multiple myeloma patients by artificial neural networks coupled with mass spectrometry of peripheral blood plasma. Sci Rep. 2019;9:7975.
Chanukuppa V, More TH, Taunk K, Taware R, Chatterjee IA, Sharma S, et al. Serum metabolomic alterations in multiple myeloma revealed by targeted and untargeted metabolomics approaches: a pilot study. Rsc Adv. 2019;9:29522–32.
Steiner N, Muller U, Hajek R, Sevcikova S, Borjan B, Johrer K, et al. The metabolomic plasma profile of myeloma patients is considerably different from healthy subjects and reveals potential new therapeutic targets. PLoS One. 2018;13:e0202045.
Veskovski L, Andersson PO, Turesson I, Malmodin D, Pedersen A, Mellqvist UH. Serum metabolomic profiling correlated with ISS and clinical outcome for multiple myeloma patients treated with high dose melphalan and autologous stem cell transplantation. Blood. 2021;97:79–88.
Fei F, Ma T, Zhou X, Zheng M, Cao B, Li J. Metabolic markers for diagnosis and risk-prediction of multiple myeloma. Life Sci. 2020;265:118852.
Gonsalves WI, Ramakrishnan V, Hitosugi T, Ghosh T, Jevremovic D, Dutta T, et al. Glutamine-derived 2-hydroxyglutarate is associated with disease progression in plasma cell malignancies. JCI Insight. 2018;3:e94543.
Wang QT, Li YZ, Liang YF, Hu CJ, Zhai YH, Zhao GF, et al. Construction of a multiple myeloma diagnostic model by magnetic bead-based MALDI-TOF mass spectrometry of serum and pattern recognition software. Anat Rec. 2009;292:604–10.
He A, Bai J, Huang C, Yang J, Zhang W, Wang J, et al. Detection of serum tumor markers in multiple myeloma using the CLINPROT system. Int J Hematol. 2012;95:668–74.
Zhang HT, Tian EB, Chen YL, Deng HT, Wang QT. Proteomic analysis for finding serum pathogenic factors and potential biomarkers in multiple myeloma. Chin Med J. 2015;128:1108–13.
Barcelo F, Gomila R, de Paul I, Gili X, Segura J, Perez-Montana A, et al. MALDI-TOF analysis of blood serum proteome can predict the presence of monoclonal gammopathy of undetermined significance. PLoS One. 2018;13:e0201793.
Ma TZ, Piao Z, Jin SY, Kwak YG. Differential expression of serum proteins in multiple myeloma. Exp Ther Med. 2019;17:649–56.
Bhattacharyya S, Epstein J, Suva LJ. Biomarkers that discriminate multiple myeloma patients with or without skeletal involvement detected using SELDI-TOF mass spectrometry and statistical and machine learning tools. Dis Markers. 2006;22:245–55.
Dowling P, Hayes C, Ting KR, Hameed A, Meiller J, Mitsiades C, et al. Identification of proteins found to be significantly altered when comparing the serum proteome from Multiple Myeloma patients with varying degrees of bone disease. Bmc Genom. 2014;15:904.
Sasser AK, Casneuf T, Amaratunga D, Tryputsen V, Kumar SK, Rajkurnar V, et al. Development of a serum biomarker panel that predicts imminent risk of multiple myeloma (MM) progression from premalignancy. Blood. 2013;122:3115.
Zingone A, Wang W, Corrigan-Cummins M, Wu SP, Plyler R, Korde N, et al. Altered cytokine and chemokine profiles in multiple myeloma and its precursor disease. Cytokine. 2014;69:294–7.
Mailankody S, Devlin SM, Korde N, Lendvai N, Lesokhin A, Landau H, et al. Proteomic profiling in plasma cell disorders: a feasibility study. Leuk Lymphoma. 2017;58:1757–9.
Bai J, Yang Y, Wang J, Zhang L, Wang F, He A. Variability of serum novel serum peptide biomarkers correlates with the disease states of multiple myeloma. Clin Proteom. 2019;16:17.
Hsieh FY, Tengstrand E, Pekol TM, Guerciolini R, Miwa G. Elucidation of potential bortezomib response markers in mutliple myeloma patients. J Pharm Biomed. 2009;49:115–22.
Rajpal R, Dowling P, Meiller J, Clarke C, Murphy WG, O’Connor R, et al. A novel panel of protein biomarkers for predicting response to thalidomide-based therapy in newly diagnosed multiple myeloma patients. Proteomics. 2011;11:1391–402.
Luczak M, Kubicki T, Rzetelska Z, Szczepaniak T, Przybylowicz-Chalecka A, Ratajczak B, et al. Comparative proteomic profiling of sera from patients with refractory multiple myeloma reveals potential biomarkers predicting response to bortezomib-based therapy. Pol Arch Intern Med. 2017;127:392–400.
Ting KR, Henry M, Meiller J, Larkin A, Clynes M, Meleady P, et al. Novel panel of protein biomarkers to predict response to bortezomib-containing induction regimens in multiple myeloma patients. BBA Clin. 2017;8:28–34.
Moloudizargari M, Abdollahi M, Asghari MH, Zimta AA, Neagoe IB, Nabavi SM. The emerging role of exosomes in multiple myeloma. Blood Rev. 2019;38:100595.
Reale A, Carmichael I, Xu R, Mithraprabhu S, Khong T, Chen M, et al. Human myeloma cell- and plasma-derived extracellular vesicles contribute to functional regulation of stromal cells. Proteomics. 2021;Feb 12:e2000119.
Manier S, Liu CJ, Avet-Loiseau H, Park J, Shi J, Campigotto F, et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood. 2017;129:2429–36.
Sedlarikova L, Bollova B, Radova L, Brozova L, Jarkovsky J, Almasi M, et al. Circulating exosomal long noncoding RNA PRINS—first findings in monoclonal gammopathies. Hematol Oncol. 2018;36:786–91.
Xu H, Han H, Song S, Yi N, Qian C, Qiu Y, et al. Exosome-transmitted PSMA3 and PSMA3-AS1 promote proteasome inhibitor resistance in multiple myeloma. Clin Cancer Res. 2019;25:1923–35.
Harshman SW, Canella A, Ciarlariello PD, Agarwal K, Branson OE, Rocci A, et al. Proteomic characterization of circulating extracellular vesicles identifies novel serum myeloma associated markers. J Proteom. 2016;136:89–98.
Kvorning SL, Nielsen MC, Andersen NF, Hokland M, Andersen MN, Moller HJ. Circulating extracellular vesicle-associated CD163 and CD206 in multiple myeloma. Eur J Haematol. 2020;104:409–19.
Reale A, Khong T, Xu R, Chen M, Mithraprabhu S, Bingham N, et al. Human plasma extracellular vesicle isolation and proteomic characterization for the optimization of liquid biopsy in multiple myeloma. In: Posch A (ed). Proteomic profiling: methods and protocols. Springer US: New York, NY, 2021;2261:151–91.
Krishnan SR, Luk F, Brown RD, Suen H, Kwan Y, Bebawy M. Isolation of human CD138(+) microparticles from the plasma of patients with multiple myeloma. Neoplasia. 2016;18:25–32.
Krishnan SR, De Rubis G, Suen H, Joshua D, Kwan YL, Bebawy M. A liquid biopsy to detect multidrug resistance and disease burden in multiple myeloma. Blood Cancer J. 2020;10:37.
Caivano A, Laurenzana I, De Luca L, La Rocca F, Simeon V, Trino S, et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumour Biol. 2015;36:9739–52.
Krishnan A, Adhikarla V, Poku EK, Palmer J, Chaudhry A, Biglang-Awa VE, et al. Identifying CD38+ cells in patients with multiple myeloma: first-in-human imaging using copper-64-labeled daratumumab. Blood Adv. 2020;4:5194–202.
Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750.
Bergin K, Moore E, McQuilten Z, Wood E, Augustson B, Blacklock H, et al. Design and development of the Australian and New Zealand (ANZ) myeloma and related diseases registry. BMC Med Res Methodol. 2016;16:151.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mithraprabhu, S., Chen, M., Savvidou, I. et al. Liquid biopsy: an evolving paradigm for the biological characterisation of plasma cell disorders. Leukemia 35, 2771–2783 (2021). https://doi.org/10.1038/s41375-021-01339-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01339-6
This article is cited by
-
Electrical detection of RNA cancer biomarkers at the single-molecule level
Scientific Reports (2023)
-
Cell-free DNA chromosome copy number variations predict outcomes in plasma cell myeloma
Blood Cancer Journal (2023)
-
Circulating tumour DNA analysis predicts relapse and improves risk stratification in primary refractory multiple myeloma
Blood Cancer Journal (2023)
-
Progress of modern imaging modalities in multiple myeloma
International Journal of Hematology (2022)
-
Monitoring multiple myeloma in the peripheral blood based on cell-free DNA and circulating plasma cells
Annals of Hematology (2022)