Abstract
Large granular lymphocyte leukemia (LGLL) is characterized by somatic gain-of-function STAT3 mutations. However, the functional effects of STAT3 mutations on primary LGLL cells have not been studied in detail. In this study, we show that CD8+ T cells isolated from STAT3 mutated LGLL patients have high protein levels of epigenetic regulators, such as DNMT1, and are characterized by global hypermethylation. Correspondingly, treatment of healthy CD8+ T cells with IL-6, IL-15, and/or MCP-1 cytokines resulted in STAT3 activation, increased DNMT1, EZH2, c-MYC, l-MYC, MAX, and NFκB levels, increased DNA methylation, and increased oxidative stress. Similar results were discovered in KAI3 NK cells overexpressing gain-of-function STAT3Y640F and STAT3G618R mutants compared to KAI3 NK cells overexpressing STAT3WT. Our results also confirm that STAT3 forms a direct complex with DNMT1, EZH2, and HDAC1. In STAT3 mutated LGLL cells, DNA methyltransferase (DNMT) inhibitor azacitidine abrogated the activation of STAT3 via restored SHP1 expression. In conclusion, STAT3 mutations cause DNA hypermethylation resulting in sensitivity to DNMT inhibitors, which could be considered as a novel treatment option for LGLL patients with resistance to standard treatments.
Similar content being viewed by others
Introduction
Large granular lymphocyte leukemia (LGLL) is a rare chronic lymphoproliferative disorder, which in over 80% of the cases results from abnormal clonal expansion of CD3 + CD8 + cytotoxic T cells [1]. Less frequently, LGLL originates from the expansion of natural killer (NK) cells (chronic lymphoproliferative disorder of NK cells, CLPD-NK). Both T-cell LGLL and CLPD-NK are chronic and indolent diseases, but instances of aggressive leukemias have also been reported [2].
Somatic gain-of-function (GOF) STAT3 mutations are a hallmark of LGLL and occur in both T and NK forms of the disease [3,4,5,6,7,8]. LGLL patients with STAT3 mutations have more often neutropenia and rheumatoid arthritis (RA) [3, 9, 10], and mutations may also confer reduced overall survival [5]. Although STAT3 activation, irrespective of mutation status, is one of the key features and a known mediator of cytokine signaling and inflammation in LGLL [1, 11,12,13], the functional consequences of STAT3 activation in LGLL remain to be fully elucidated.
Cytokines induce differentiation and proliferation of lymphocytes, and aberrant cytokine signaling can be observed in many inflammatory conditions [14]. Interleukin-6 (IL-6) is elevated in RA leading to JAK/STAT3 signaling activation, and IL-6 and IL-6 receptor inhibitors are used in RA treatment [15]. In addition to the well-established role in chronic inflammatory diseases, cytokine dysregulation may also drive tumor initiation and progression [16]. In LGLL mouse model, excessive IL-15 induced DNMT3b upregulation and global DNA hypermethylation, and IL-15 may drive LGLL formation [17,18,19].
As the role of STAT3 in regulating DNA methylation in LGLL is currently unknown, we investigated whether inflammatory cytokine stimulation and STAT3 activation modify the epigenetic machinery in CD8+ T cells and could thereby affect disease pathogenesis in LGLL. Using LGLL patients samples, healthy human CD8+ T cells, and KAI3 NK cell line, we show that cytokine stimulation or overexpression of GOF STAT3 variants lead to increased protein levels of epigenetic regulators, increased global DNA methylation levels, and increased reactive oxygen species (ROS) production. Treatment with DNMT inhibitor abrogated STAT3 mediated changes in SHP1 dependent manner, paving the way for novel targeted therapeutic approaches.
Materials and methods
Materials and methods
More detailed descriptions of the methods and analyses are available in the Supplementary Methods.
Study participants
Study participants were recruited at Helsinki University Hospital (HUH), Helsinki, Finland. Healthy controls were collected from Finnish Red Cross Blood Service (Helsinki, Finland). All participants gave written informed consent before participation. The ethics committee in the HUH approved the study, and it was conducted in accordance with the Declaration of Helsinki. Patients’ clinical characteristics are shown in Supplementary Table 1 as well as assays in which samples were used.
Cell line
Previously published [20] KAI3 NK cells engineered to express STAT3 wildtype (STAT3WT) or activating mutations of STAT3 (STAT3G618R and STAT3Y640F) were used to validate the functional effects of STAT3 mutations. Cells were maintained in RPMI 1640 Medium containing 10% FBS, 1% penicillin/streptomycin, 2 mM L-glutamine, and 100 IU/ml recombinant human Interleukin-2. Mycoplasma test was performed using PCR Mycoplasma Test kit (Cat. PK-CA91-1096, PromoCell, Heidelberg, Germany). Cells were authenticated using GenePrint10 (Promega, Madison, WA, USA). The DSMZ-STR, ATCC-STR, JCRB-STR, and ICLC-STR databases were used to compare the results.
Plasma cytokine measurements
Plasma cytokine measurement was performed using a Proseek Multiplex Inflammation panel by Olink Proteomics (Uppsala, Sweden) [21]. The relative protein levels are expressed as Normalized Protein eXpression (NPX) in 2-log scale. A larger NPX level indicates a higher protein expression in the sample.
Human primary CD8+ T cells sorting
Mononuclear cells (MNCs) were isolated using a Ficoll gradient centrifuge. Primary CD8+ T cells were sorted by autoMACS® Pro Separator (Miltenyi Biotec, USA) with CD8 MicroBeads (Miltenyi Biotec, Cat. 130-045-201) from isolated MNCs. The final pools contained >95% purified cells.
RNA sequencing
Total RNA was extracted from PBMCs of T-LGLL patients and healthy controls. RNA-sequencing using Illumina Hiseq 2000 and further bioinformatic processing were performed as previously reported [22].
siRNA transfection
siRNAs were purchased from Dharmacon (Chicago, IL, USA). Cells were transfected with 50 nmol/L siRNA using Lipofectamine™ 2000 Transfection Reagent (Thermo Fisher Scientific) in 12-well plates according to manufacturer’s protocols. siRNA sequences are described in Supplementary Table 2.
Drug sensitivity and resistance testing
Drug sensitivity and resistance testing were performed with primary CD8+ T cells of both T-LGLL patients harboring STAT3Y640F mutation and healthy controls. Azacitidine (HY-10586, CAS 320-67-2) was purchased from MedChemExpress (Monmouth Junction, NJ, USA). The experiment was performed as previously described [23].
Data analysis and statistics
Statistical analysis was performed using GraphPad Prism (Version 8.4.3) and R 4.0.3. Comparisons between the two groups were made using unpaired two side t-test and Mann–Whitney U test. P value < 0.05 was considered as statistically significant.
Results
Plasma inflammatory cytokine levels are increased in LGLL patients
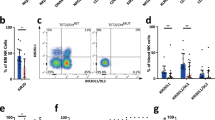
We measured plasma cytokine levels using Olink inflammation assay panel [21], including 91 proteins in nine T-LGLL patients (STAT3Y640F, STAT3D661Y, STAT3Y657ins, and without STAT3 mutations) and eight healthy controls. 34 out of 91 cytokines were higher in T-LGLL patients compared to healthy controls (Fig. 1A, Supplementary Table 3), suggesting broad cytokine upregulation. Specifically, IL-6 and IL-15RA were higher in T-LGLL patients compared to healthy controls irrespective of STAT3 mutation status (Fig. 1A). Of note, IL-15 was not included in the Olink inflammation panel used (Supplementary Table 3). Monocyte chemoattractant protein 1 (MCP-1) levels were also upregulated in patients with STAT3 mutations, whereas for example IL-2 and IL-4 levels were similar between patients and healthy controls (Fig. 1A).
A Plasma cytokine levels were measured using a Proseek Multiplex Inflammation I immunoassay (Olink Proteomics, Sweden) in CD8+ T cells of healthy controls (HC, n = 8), LGLL patients harboring STAT3 somatic mutation (n = 3, STAT3Y640F; n = 1, STAT3D661Y; n = 1, STAT3Y657ins), or without STAT3 mutations (n = 4). Left, A volcano plot of the normalized protein expression (NPX). The plot shows 91 plasma cytokines differentially expressed between CD8+ T cells from LGLL patients and healthy controls. The red dots indicate significantly higher genes in LGLL patients or healthy controls. Solid line indicates P value < 0.05 (Mann–Whitney U test). Right, plasma protein NPX level of IL-6, IL-15RA, MCP-1, IL-2, and IL-4. Helthy control vs LGLL with STAT3 GOF mutations vs LGLL without STAT3 mutations. B Relative mRNA expression of IL-6, IL-15, and MCP-1 were determined using RT-qPCR in the CD8+ T cells of healthy controls (HC), T-LGLL patients harboring STAT3 somatic mutation (T-LGLL STAT3 mut; n = 5, STAT3Y640F; n = 1, STAT3D661Y), and without STAT3 mutations (T-LGLL no mut) (n = 6 for each group). The sequences of primers were listed in Supplementary Table 4. C RNA-seq analysis of IL-6, IL-15, and MCP-1 in CD8+ T cells of T-LGLL patients harboring STAT3 mutations (T-LGLL STAT3 mut; n = 5, STAT3Y640F; n = 5, STAT3D661Y), patients without STAT3 mutations (T-LGLL no mut, n = 3) and healthy controls (HC, n = 5). Each dot represents one individual. Data are expressed as mean ± SD, and statistically significant difference was evaluated using Mann–Whitney U test. *P < 0.05; **P < 0.01.
We next examined IL-6, IL-15, and MCP-1 mRNA expression levels in CD8+ T cells using RT-qPCR. IL-15 expression levels were significantly higher in STAT3 mutated T-LGLL, whereas only borderline differences in expression levels were observed for IL-6 and MCP-1 (Fig. 1B), suggesting that other cell types contribute to circulating cytokine levels [13]. Similar expression levels were also seen in RNA-sequencing of normal and T-LGLL CD8+ T cells (STAT3Y640F, STAT3D661Y, and without STAT3 mutations) (Fig. 1C, Supplementary Fig. 1).
Epigenetic regulator levels are linked with STAT3 mutation status in T-LGLL
Using nuclear fractions of CD8+ T cells, we observed high STAT3 phosphorylation levels in T-LGLL patients harboring STAT3Y640F mutation, in accordance with previous reports (Fig. 2A) [3, 4, 24]. In addition, total STAT3 levels were significantly increased in T-LGLL patients compared with healthy controls.
Western blot analysis of CD8+ T cells from T-LGLL patients. Nuclear fractions from CD8+ T cells of healthy controls (HC, n = 5), T-LGLL patients harboring STAT3Y640F mutation (STAT3Y640F, n = 5) and without STAT3 mutations (No mut, n = 5) were extracted. A Left, data are shown as phosphorylated STAT3 (p-STAT3) and STAT3. Right, quantitative presentation of Western blot assays using Image J (Version 2.0.0). Protein levels were first normalized to LAMINB1, a loading control. The p-STAT3 level was normalized by dividing intensity of p-STAT3 by total STAT3. B Left, DNMT1, DNMT3B, and EZH2 protein expression levels. Right, quantitative presentation of the protein levels. DNMT1, DNMT3B, and EZH2 protein levels were normalized with LAMINB1. Each dot represents the protein expression level from one individual. Error bar present mean ± SD (n = 5 per group), and statistically significant difference was evaluated using Mann–Whitney U test. *P < 0.05; ***P < 0.001; ****P < 0.0001. C KAI3 NK cells harboring STAT3WT (WT), STAT3G618R mutation (G618R) and STAT3Y640F mutation (Y640F) were serum starved for 12 h followed by nucleus extraction. Protein samples were analyzed via Western blot analysis using the indicated antibodies. Data are representative of three independent experiments. Protein quantification is presented in Supplemental Fig. 3A. D KAI3 NK cells (STAT3WT, STAT3G618R, and STAT3Y640F) were transfected with STAT3-siRNA (si-STAT3) and control siRNA (si-con) for 72 h followed by serum starvation for 12 h Western blot analysis was performed with p-STAT3 and STAT3 specific antibodies. LAMINB1 served as loading control and data are representative of three independent experiments. E Cell proliferation assay was peformed using STAT3 siRNA-mediated knockdown in KAI3 NK cells. KAI3 NK cells stably expressing STAT3WT and gain-of-function STAT3 variants (STAT3G618R and STAT3Y640F) were cultured with STAT3-siRNA (si-STAT3) and control siRNA (si-con) for 48 h The cells (10,000 cells/well) were incubated in RPMI1640 with reduced IL-2 (25IU/ml) with extra si-RNAs for indicated days, and the cell proliferation was measured by DNA fluorescence-based assay. The cell numbers were calculated with a standard curve based on the fluorescence intensity. Error bar present mean ± SD of three independent experiments, and statistically significant difference was evaluated using unpaired t-test (STAT3Y640F si-con vs STAT3Y640F si-STAT3, STAT3G618R si-con vs STAT3G618R si-STAT3). *P < 0.05; **P < 0.01; ***P < 0.001.
To examine whether STAT3 regulates DNMT activity and DNA methylation in T-LGLL, we quantified DNMT protein levels in patient samples. Interestingly, DNMT1 and DNMT3B levels were significantly higher in CD8+ T cells of the LGLL patients with STAT3Y640F GOF mutation compared to healthy controls (Fig. 2B). The protein level of EZH2, a histone H3K27 methyltransferase, was also higher in T-LGLL patients, particularly in patients with STAT3Y640F mutation. In addition, we observed high level of MYC protein in T-LGLL patients harboring STAT3Y640F GOF mutation (Supplementary Fig. 2).
To further investigate whether STAT3Y640F mutation can upregulate epigenetic regulators, we used KAI3 NK cells previously engineered to overexpress exogenous STAT3 wildtype (STAT3WT) and hyperactive STAT3 variant (STAT3Y640F) [20] found in T-LGLL patients [3,4,5]. We also tested KAI3 cells expressing STAT3G618R variant [20] to further examine the effect of STAT3 GOF mutations. DNMT1 and DNMT3B were significantly upregulated in KAI3 NK cells overexpressing STAT3 GOF variants compared to cells overexpressing STAT3WT (Fig. 2C, Supplementary Fig. 3A). In addition, EZH2 protein level and levels of H3K27me3, the end-product of histone methylation catalyzed by EZH2 as part of the PRC2 complex [25], were increased in cells overexpressing STAT3 GOF variants (Fig. 2C, Supplementary Fig. 3A). DNMT3A protein was not detected in KAI3 NK cells (Supplementary Fig. 3B). Similar to patient cells, the expression of MYC family proteins, including C-MYC and L-MYC, were increased in KAI3 NK cells overexpressing STAT3 GOF mutations (Fig. 2C). Further, MYC-associated factor X (MAX) [26] was upregulated in KAI3 NK cells expressing STAT3 variants (Fig. 2C). The protein level of p65, a subunit of NF-κB reported to be upregulated in MYC-dependent manner upon IL-15 stimulation [17], was also increased in hyperactive STAT3 KAI3 NK cells (Fig. 2C). Although HDAC1 has been reported to be upregulated by excessive production of IL-15 in T cells [27], we did not find any difference in HDAC1 levels according to the STAT3 status (Fig. 2C, Supplementary Fig. 3A).
Overexpression of GOF STAT3 variants have been reported to promote cell proliferation in KAI3 NK cells under limiting IL-2 concentrations [20]. To further validate that this was due to STAT3 activation, we performed si-RNA mediated STAT3 knockdown in KAI3 NK cells (Fig. 2D). While STAT3 GOF expression led to increased cell proliferation in KAI3 NK cells, STAT3 knockdown rescued this phenotype (Fig. 2E). We additionally confirmed that STAT3 knockdown attenuated C-MYC (Supplementary Fig. 4), further supporting a link between STAT3 and MYC pathway.
Cytokines activate STAT3 and epigenetic regulator signaling in healthy CD8+ T cells
To address whether high cytokine levels are sufficient to activate STAT3 and the epigenetic machinery in CD8+ T cells, we isolated primary CD8+ T cells from healthy blood donors. These cells were cultured with IL-6, IL-15, and MCP-1, which were selected based on their higher levels in T-LGLL patients relative to healthy controls (Fig. 1A). Cytokines increased CD8+ T cell proliferation rate after 12 h of stimulation (Supplementary Fig. 5). Using four different cytokine stimulation conditions (IL-6, IL15, MCP-1, IL15 plus MCP-1) for equal time span in saturating cytokine concentrations, we observed that each cytokine increased the phosphorylation of STAT3, as well as total protein levels of STAT3, DNMT1, DNMT3B, EZH2, and MYC family proteins (Fig. 3A, Supplementary Fig. 6A). DNMT3A was not detected (Supplementary Fig. 6B). CD8+ T cells treated with a combination of IL-15 and MCP-1 showed the highest level of p-STAT3 (Fig. 3A), suggesting that in addition to direct effects of STAT3 mutations, increased levels of IL-6, IL-15, or MCP-1 (Fig. 1A, B) may further hyperactivate STAT3, DNMT1, and MYC signaling in LGLL.
A Western blot analysis of CD8+ T cells of healthy controls. Cells were cultured with IL-6, IL-15, MCP-1, IL-15 plus MCP-1 and without cytokine (con) in RPMI1640 medium for 12 h (100 ng/ml for each cytokine). Nuclear fractions of the cells were used to detect the indicated proteins. LAMINB1 was used as loading control. B–F Physical interaction of STAT3 with DNMT1, EZH2, and HDAC1. CD8+ T cells of healthy controls were cultured with IL-6, IL-15, MCP-1, and IL15 plus MCP-1 in RPMI1640 medium for 12 h to induce STAT3 activation. Co-Immunoprecipitation was performed with the nuclear protein lysates of CD8+ T cells. B The nuclear extracts (10%) of the cells were used as input control. Co-IP was performed with antibody against C STAT3, D DNMT1, E HDAC1, and F EZH2. Rabbit IgG Isotype control (IgG) was used as a negative control for non-specific binding for Co-IP. Immunoprecipitated complexes were probed using the antibodies as indicated. Data are representative of three independent experiments. G Simplified scheme of the complex of STAT3, DNMT1, EZH2, and HDAC1 using BioRender (Toronto, Canada).
STAT3 directly binds epigenetic regulator proteins
To understand the connection between STAT3 and epigenetic modifiers, we performed Co-immunoprecipitation (Co-IP) in CD8+ T cells isolated from healthy controls and stimulated with IL-6, IL-15, MCP-1, or IL-15 plus MCP-1. STAT3 was able to bind DNMT1, EZH2, and HDAC1 (Fig. 3B–E). EZH2 directly interacted with STAT3 but not within a higher-order complex including HDAC1 or DNMT1 (Fig. 3F–G). Consistent with findings in stimulated healthy CD8+ T cells, we observed stronger interaction between STAT3 and DNMT1 as well as between STAT3 and HDAC1 in KAI3 NK cells overexpressing GOF STAT3 compared to STAT3WT (Supplementary Fig. 7).
STAT3 activation is associated with DNA hypermethylation
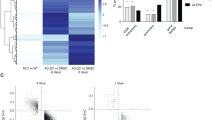
We next quantified global DNA methylation in healthy and T-LGLL CD8+ T cells as well as in KAI3 NK cells. CD8+ T cells from T-LGLL patients harboring STAT3Y640F mutation had the highest global DNA methylation levels (median = 1.44) compared to cells from T-LGLL patients without STAT3 mutations (median = 0.45) or from healthy controls (median = 0.365) (Fig. 4A). Similarly, the methylation levels were significantly higher in STAT3Y640F (median = 1.90) and STAT3G618R (median = 1.72) cells compared to STAT3WT KAI3 NK cells (median = 0.86) (Fig. 4B). Furthermore, stimulation of healthy control CD8+ T cells with cytokines led to higher global DNA methylation levels (Fig. 4C). IL-15 (median = 0.70) showed the highest impact on upregulating DNA methylation among the cytokine conditions used (median: IL-6 = 0.64, MCP-1 = 0.61, IL-15+MCP-1 = 0.68, without treatment = 0.40).
A Global DNA methylation levels in CD8+ T cells of healthy controls (HC, n = 6), T-LGLL patients harboring STAT3Y640F mutation (STAT3Y640F, n = 5), and T-LGLL patients without STAT3 mutations (No mut, n = 9). %5-mC were evaluated by fluorescence according to manufacturer’s instruction. B KAI3 NK cells stably expressing STAT3WT (WT), STAT3G618R (G618R) and STAT3Y640F (Y640F) were cultured without serum for 12 h followed by genomic DNA extraction and global DNA methylation measurement. C Global DNA methylation levels in CD8+ T cells of healthy controls stimulated with IL-6, IL-15, and/or MCP-1 for 12 h were measured (n = 6 for each group). Error bar present mean ± SD and statistically significant difference was evaluated using Mann–Whitney U test. *P < 0.05; **P < 0.001; ***P < 0.001. D Protein expression in KAI3 NK cells transfected with STAT3-siRNA (si-STAT3) and control siRNA (si-con) for 72 h in RPMI1640 with reduced IL-2 (25IU/ml) followed by serum starvation for 12 h. Protein extracts from siRNA-treated cells were subjected to Western blot analysis with p-STAT3, STAT3, and DNMT1 specific antibodies. LAMINB1 served as loading control and data are representative of three independent experiments. E Global methylation level in KAI3 NK cells transfected with STAT3-siRNA and con-siRNA for 72 h in RPMI1640 with reduced IL-2 (25IU/ml). Error bar present mean ± SD (n = 3 per group), and statistically significant difference was evaluated using Mann–Whitney U test. Error bar present mean ± SD and statistically significant difference was evaluated using Mann–Whitney U test. ***P < 0.001. F Simplified scheme of the STAT3 GOF mutations. Pathway components discovered to be upregulated with functional assays are marked with arrows. The schematic was made using BioRender (Toronto, Canada).
We then evaluated whether STAT3 inhibition can suppress DNA methylation using KAI3 NK cells and a small interfering RNA (siRNA) targeting STAT3. Consistent with results from a prior study [28], DNMT1 levels decreased in response to STAT3 inhibition (Fig. 4D). Global methylation levels decreased with STAT3 siRNA in KAI3 NK cells overexpressing STAT3 mutants, but not in STAT3WT cells (Fig. 4E), suggesting that the upregulated DNA methylation is caused by STAT3 activity and DNMT1 (Fig. 4E, F).
Increased ROS levels associated with STAT3 activation and cytokine stimulation
Since a link between DNA methylation and oxidative stress has been suggested [29], we measured ROS levels in CD8+ T cells from T-LGLL patients using an in vitro ROS/RNS (reactive nitrogen species) assay. ROS/RNS levels were significantly higher in T-LGLL patients harboring STAT3Y640F mutation (median = 3070) compared with both healthy controls (median = 1528) and the patients without STAT3 mutations (median = 2117) (Fig. 5A). Interestingly, KAI3 NK cells overexpressing STAT3 (either wildtype or STAT3 GOF) showed increased levels of both ROS and superoxide compared to the control KAI3 cells. Moreover, ROS levels in cells carrying STAT3 GOF mutations were higher compared to KAI3 NK cells expressing STAT3WT (Fig. 5B).
A The level of ROS/RNS in CD8+ T cells of T-LGLL patients harboring STAT3Y640F mutation (STAT3Y640F) and without STAT3 mutations (No mut) as well as healthy controls (HC) (n = 3). Error bars expressed as mean ± SD, and statistically significant difference was evaluated using unpaired t-test. *P < 0.05. B Flow cytometry analysis to examine the expression of total ROS and superoxide in KAI3 NK cells harboring STAT3WT, STAT3G618R, STAT3Y640F mutation and control KAI3 NK cells (Mock). Cells were cultured in RPMI1640 with reduced IL-2 (25IU/ml) for 72 h Then, the cells were labeled with ROS-fluorescent dye (Ex/Em= 650/675 nm) and superoxide-fluorescent dye (Ex/Em = 520/605 nm), and then analyzed using flow cytometer. KAI3 NK cells expressing empty vector (Mock) were used as a control. C Flow cytometry analysis to examine the expression of total ROS and superoxide in untreated CD8+ T cells (con) and those incubated with IL-6, IL-15, MCP-1, or IL15 plus MCP-1. CD8+ T cells were incubated with the indicated cytokines (100 ng/ml) for 12 h in RPMI1640. Cells were labeled with ROS-fluorescent dye and superoxide-fluorescent dye, and then analyzed using flow cytometer. D p-ATM, ATM, γH2AX, H2AX, p65 and p-p65 protein expression in healthy CD8+ T cells stimulated with IL-6, IL-15, and/or MCP-1 for 12 h. Quantification is presented in Supplementary Fig. 8A. E p-ATM, ATM, γH2AX, H2AX, p65, and p-p65 protein expression in CD8+ T cells of T-LGLL patients harboring STAT3Y640F mutation (STAT3Y640F) and without STAT3 mutations (No mut) as well as healthy controls (HC). Quantification is presented in Supplementary Fig. 8B.
We investigated whether cytokines can also promote ROS production in healthy CD8+ T cells. In line with our primary LGLL patient data, total ROS was significantly increased from 0.023% (control) to 9.6% (IL-6), 20.8% (IL-15), 7.2% (MCP-1), and 30.0% (IL-15 plus MCP1) (Fig. 5C). Superoxide levels were also increased in response to IL-15 (6.1%), MCP-1 (5.4%), and IL-15 plus MCP-1 (8.3%) compared to control cells (4.7%), whereas IL-6 (4.3%) had no effect on the production of superoxide. Cytokine stimulation also increased p-ATM and γH2AX in CD8+ T cells (Fig. 5D, Supplementary Fig. 8A), indicating concomitant activation of the DNA damage response pathway [30]. Ser-276 phosphorylation of NF-κB p65 was also noted in cytokine-stimulated CD8+ T cells (Fig. 5D, Supplementary Fig. 8A), suggesting ROS-dependent NF-κB signaling activation [31]. In addition, these proteins were upregulated in CD8+ T cells of T-LGLL patients harboring STAT3 GOF mutation (Fig. 5E, Supplementary Fig. 8B). Taken together, these results suggest that STAT3 driver mutations and higher p-STAT3 activation are associated with higher ROS production and activation of the DNA damage response and NF-κB pathways, potentially contributing to LGLL phenotype.
Azacitidine abrogates STAT3 activation, DNA methylation, and ROS production
To explore whether DNA hypermethylation associated with STAT3 activation could be therapeutically targeted, we evaluated the effects of azacitidine, a hypomethylating agent widely used to treat myeloid malignancies. Ex vivo drug sensitivity testing showed that T-LGLL CD8+ T cells were more sensitive to azacitidine at higher concentrations compared with CD8+ T cells from healthy controls (Fig. 6A). Next, we investigated whether azacitidine decreases the protein levels of STAT3, p-STAT3, DNMT1, and EZH2. CD8+ T cells of a T-LGLL patient harboring STAT3Y640F mutation were cultured in the presence or absence of azacitidine (1 and 10 μM) for 24, 48, and 72 h. Azacitidine inhibited p-STAT3, total STAT3, and DNMT1 in dose- and time-dependent manner (Fig. 6B, Supplementary Fig. 9). EZH2 levels were not affected by azacitidine. To validate these results, we treated KAI3 NK cells with azacitidine. While no statistically significant differences in cell viability were observed between cells overexpressing STAT3WT or GOF STAT3 variants (Fig. 6C), protein levels of pSTAT3, total STAT3, and DNMT1 were decreased with azacitidine treatment (Fig. 6D), consistent with results observed in CD8+ T cells. EZH2 level was also decreased after 72 h of 10 μM of azacitidine treatment in KAI3 NK cells (Fig. 6D, Supplementary Fig. 10).
A Drug sensitivity testing was performed on CD8+ T cells from T-LGLL patients harboring STAT3Y640F mutation (STAT3Y640F, n = 3) and healthy controls (n = 3). Cell viability inhibition by correlation of drug sensitivity score (DSS) was measured with CellTiter-Glo2.0 (Promega, USA). DSS is a quantitative measurement of a drug response based on the area under the curve (AUC) with further normalization. Higher DSS denote better killing activity. Dose-response curve of LGLL patients (red line) and healthy controls (gray line) for azacitidine. B CD8+ T cells from a T-LGLL patient harboring STAT3Y640F mutation were cultured in the presence of azacitidine (1 or 10 μM) or absence (DMSO alone) for 24, 48, and 72 h. Cell lysates were immunoblotted with antibodies against p-STAT3, STAT3, DNMT1, and EZH2. LAMINB1, a loading control. Data are representative of three T-LGLL patients harboring STAT3Y640F and quantification is presented in Supplementary Fig. 9. C Dose-response curve of KAI3 NK cells stably expressing STAT3WT (gray line), STAT3G618R (blue line), STAT3Y640F (red line) for azacitidine (n = 2 per group). DSS scores are presented in Supplementary Table 5. D KAI3 NK cells stably expressing STAT3WT, STAT3G618R and STAT3Y640F were cultured in the presence of azacitidine (1 or 10 μM) or absence (DMSO alone) for 24, 48, and 72 h in RPMI1640 with reduced IL-2 (25IU/ml). Cell lysates were immunoblotted with antibodies against p-STAT3, STAT3, DNMT1, and EZH2. LAMINB1, a loading control. Data are representative of two independent experiments. Quantification of the proteins is presented in Supplementary Fig. 10. E Global DNA methylation levels in KAI3 NK cells of STAT3WT, STAT3G618R and STAT3Y640F (for each group, n = 3) treated with 1 μM of azacitidine (upper) and 10 μM of azicitidine (lower) at 24, 48, and 72 h in RPMI1640 with reduced IL-2 (25IU/ml). %5-mC were evaluated by fluorescence according to manufacturer’s instruction. Error bar present mean ± SD (n = 3 per group), and statistically significant difference was evaluated using Mann–Whitney U test. **P < 0.01; ***P < 0.001; ****P < 0.0001. F ROS expression in KAI3 NK cells stably expressing STAT3WT, STAT3G618R, and STAT3Y640F. Cells were cultured in the presence of azacitidine (1 or 10 μM) or absence (DMSO alone) for 72 h in RPMI1640 with reduced IL-2 (25IU/ml). Cells were labeled with ROS-fluorescent dye then analyzed using flow cytometer. Error bars expressed as mean ± SD, and statistically significant difference was evaluated using unpaired T-test. **P < 0.01; ***P < 0.001. The representative of three independent experiments is presented in Supplementary Fig. 12A. G p-p65 and p65 protein expression level from azacitidine (1 or 10 μM) treated CD8+ T cells of T-LGLL patient harboring STAT3Y640F mutation. Data are representative of three T-LGLL patients harboring STAT3Y640F. H p-p65 and p65 protein expression level from azacitidine (1 or 10 μM) treated KAI3 NK cells stably expressing STAT3WT, STAT3G618R, and STAT3Y640F. Data are representative of two independent experiments.
In line with the hypomethylating mechanism of action of azacitidine, KAI3 NK cells showed decreased global methylation levels in time and dose-dependent manner after azacitidine treatment (Fig. 6E, Supplementary Fig. 11). ROS levels were decreased in KAI3 NK cells expressing either STAT3WT and STAT3 GOF variants (Fig. 6F, Supplementary Fig. 12A). In addition, azacitidine reduced Ser-276 phosphorylation of p65 in CD8+ T cells of LGLL patients (Fig. 6G) and KAI3 NK cells (Fig. 6H). Given that ROS can stimulate overall tyrosine phosphorylation-dependent signaling [32], we additionally assessed the effect of azacitidine on total phosphotyrosine levels in KAI3 NK cells. Azacitidine significantly reduced whole cell tyrosine-phosphorylated proteins (25–260 kDa), including p-STAT3 (Supplementary Fig. 12B). Together, these results suggest that DNMT inhibitors can influence epigenetic programming and ROS production in STAT3 activation-dependent manner, with potential clinical applicability in LGLL.
Azacitidine inhibits STAT3 activation by SHP1 restoration
Previous studies have shown that SHP1 negatively regulates JAK/STAT3 signaling pathway [33, 34]. Basal expression of SHP1 was lower in CD8+ T cells of T-LGLL patients harboring STAT3Y640F mutations compared to both healthy controls and T-LGLL patients without STAT3 mutations (Fig. 7A, Supplementary Fig. 13). Furthermore, azacitidine treatment led to increased SHP1 levels in CD8+ T cells isolated from T-LGLL patients (Fig. 7B, Supplementary Fig. 14). SHP1 levels were lower in KAI3 NK cells overexpressing GOF STAT3 mutations compared to STAT3WT cells at baseline; however, azacitidine treatment increased SHP1 levels in GOF STAT3 cells (Fig. 7C). Next, KAI3 NK cells were transfected with control and SHP1 siRNA for 48 h followed by azacitidine treatment. Knockdown of SHP1 upregulated p-STAT3 and DNMT1 in STAT3WT cells at baseline (Fig. 7D). In addition, SHP1 knockdown attenuated p-STAT3 and DNMT1 downregulation during azacitidine treatment in KAI3 NK cells. SHP1 knockdown promoted cell proliferation in STAT3WT but not in GOF STAT3 cells, providing further support for the link between STAT3 upregulation and cell proliferation (Fig. 7E). We also confirmed that siRNA-mediated STAT3 knockdown restored SHP1 expression (Fig. 7F). Together, these findings suggest that SHP1 controls STAT3 activity and inhibition of STAT3 during azacitidine treatment (Fig. 7G).
A SHP1 protein levels at baseline in CD8+ T cells of healthy controls (HC, n = 3), T-LGLL patients harboring STAT3Y640F mutation (STAT3Y640F, n = 3) and T-LGLL patient without STAT3 mutations (No mut, n = 3). Quantification is presented in Supplementary Fig. 13. B SHP1 protein levels in CD8+ T cells of a T-LGLL patient harboring STAT3Y640F mutation treated with azacitidine (1 or 10 μM) for 24, 48, and 72 h Data are representative of three T-LGLL patients harboring STAT3Y640F and quantification is presented in Supplementary Fig. 14. C SHP1 expression in KAI3 NK cells (STAT3WT, STAT3G618R, and STAT3Y640F) treated with azacitidine (1 or 10 μM) for 24, 48, and 72 h in RPMI1640 with reduced IL-2 (25IU/ml) followed by Western blot assay. D KAI3 NK cells stably expressing STAT3WT, STAT3G618R, and STAT3Y640F were treated with SHP1-siRNA (si-SHP1) and control siRNA (si-con) for 72 h followed by azacitidine treatment for 72 h in RPMI1640 including reduced amount of IL-2 (25IU/ml). Western blot was performed with the use of anti-pSTAT3, anti-STAT3, anti-DNMT1, anti-SHP1, and anti-LaminB1 antibodies. E KAI3 NK cells stably expressing STAT3WT and GOF STAT3 variants (STAT3G618R and STAT3Y640F) were cultured with SHP1-siRNA (si-SHP1) and control siRNA (si-con) for 48 h. Next, siRNAs transfected cells (10,000 cells/well) were cultured in RPMI1640 with reduced IL-2 (25IU/ml) with extra siRNAs for 72 h, and the cell proliferation was measured by DNA fluorescence-based assay. Cell numbers were calculated with a standard curve based on the fluorescence intensity. Error bar present mean ± SD of three independent experiments, and statistically significant difference was evaluated using unpaired t-test (STAT3WT si-con vs STAT3WT si-SHP1). *P < 0.05; **P < 0.01. F SHP1 expression in KAI3 NK cells transfected with STAT3-siRNA (si-STAT3) and control siRNA (si-con) for 72 h. The cells were serum starved for 12 h followed by Western blot assay. G Simplified scheme of the effect of azacitidine. Pathway components discovered to be upregulated or downregulated with functional assays are marked with arrows. The scheme was made using BioRender (Toronto, Canada).
Discussion
Activating STAT3 somatic mutations can be found both in patients with T-cell LGLL (40-50% of cases with CD8 + CD3 + phenotype) and in CLPD-NK (20-30% of patients) [3, 6]. STAT3 is also mutated in other T and NK cell neoplasias such as mature T cell lymphomas and aggressive NK cell leukemias [35,36,37,38]. Interestingly, germline STAT3 GOF variants have also been associated with early LGL lymphoproliferation [39, 40]. Furthermore, a role for JAK/STAT activation in LGLL pathogenesis even in patients without STAT3 mutations has been suggested [1, 6, 12, 13]. Nevertheless, the consequences of STAT3 activation in pathological T- and NK-cell proliferations remain elusive. In our study, we show that in primary LGLL cells GOF STAT3 mutations alter autoregulation of p-STAT3 and drive changes in epigenetic regulator levels, global DNA hypermethylation, and ROS production. In addition to LGLL patient cells, we used KAI NK cells and cytokine-stimulated CD8+ T cells to validate the consequences of STAT3 pathway activation although it needs to be taken into account that due to gain of function STAT3 mutations, the STAT3 activation in T-LGLL is chronic compared to transient changes induced by cytokine stimulation. Our results were well recapitulated in these models, and we observed higher-order protein complex interactions between STAT3 and EZH2, HDAC1, and DNMT1 in both healthy CD8+ T cells with proinflammatory cytokine treatment and KAI3 NK cells overexpressing GOF STAT3 variants, indicating a direct link between STAT3 and epigenetic regulation. Azacitidine, a hypomethylating agent, rescued these phenotypes in SHP1 dependent manner. Together, our results suggest a central role for STAT3 and epigenetic changes to drive LGLL pathogenesis, and provide rationale for testing hypomethylating agents in the treatment of LGLL patients resistant to standard therapies.
Prior studies have shown elevated levels of circulating inflammatory cytokines in LGLL [13, 41,42,43]. The exact mechanism and dominance of certain cytokines versus complex action of several cytokines remains largely enigmatic. We confirmed increased cytokine levels in T-LGLL patients compared to healthy controls. Increased global DNA methylation pattern was found both in CD8 + LGLL cells and in response to stimulation with IL-6, IL-15, and MCP-1 in healthy CD8+ T cells. These global epigenetic changes may partly confer a selection pressure on T cell clones, facilitating clonal expansion after cytokine stimulation. While the increased total methylation pattern in response to cytokine stimulation in CD8+ T cells was confirmed in the fluorescence-based assay, DNA methylation sequencing analyses are warranted to explore the methylated genomic sites in detail. In addition, further studies using primary LGLL cells could link alterations in cytokine profiles and global DNA methylation with key gene expression profiles.
We observed higher levels of circulating IL-15RA in T-LGLL patients samples, and IL-15 promoted epigenetic changes in healthy CD8+ T cells. IL-15-producing macrophages and dendritic cells have been reported to cross-represent IL-15 to promote proliferation and cytotoxicity in LGL [19]. IL-15 can initiate LGLL in transgenic mice via the induction of MYC, p65, and HDAC1 with DNMT3B overexpression and DNA hypermethylation [17]. In addition, IL-15 can maintain the survival of leukemic clones in T-LGL and other T malignancies [44]. We also observed the activation of these proteins in healthy CD8+ T cells stimulated with cytokines, and validated these findings in KAI3 NK cells overexpressing GOF STAT3 mutations, indicating that enhanced p-STAT3 may promote LGLL pathogenesis in part via induction of these oncogenic pathways by cytokine stimuli.
STAT3 interacts with many key epigenetic regulators and other transcription factors, coactivators, and corepressors to reprogram transcription [45,46,47]. DNMT1 is closely associated with STAT3 signaling in malignant T cells [48], and our results suggest that STAT3 directly binds DNMT1, EZH2, and HDAC1 to regulate DNA methylation. We showed that STAT3 interacts with DNMT1 and HDAC1 both in CD8+ T cells and KAI3 NK cells, which is in line with previous studies showing STAT3-DNMT1-HDAC1 complex in lymphoma cells [49]. These complexes may contribute to the link between p-STAT3 activation and global DNA methylation.
ROS levels were higher in CD8+ T cells of LGLL patients compared to healthy controls, which is consistent with previous studies showing increased level of ROS in hematological malignancies [50, 51]. KAI3 NK cells expressing STAT3 GOF variants had higher levels of ROS compared to cells overexpressing STAT3WT, suggesting that ROS is associated with STAT3 activation. We also observed increased ROS levels after 12-hour cytokine stimulation in healthy CD8+ T cells. Furthermore, we found increased activation of both DNA damage and NF-κB pathways in STAT3 mutated CD8+ T cells of LGLL patients and healthy CD8+ T cells after cytokine stimulation. Rapidly accumulating DNA damage upon stimulation of CD8+ T cells may lead to increased mutagenesis and clonal selection, while chronic elevation in ROS and DNA damage may be markers of increased clonal turnover in T-LGLL patients. Given that inflammatory cytokines are commonly associated with chronic inflammation and ROS production [52], our results suggest that STAT3 activation, high ROS levels, DNA damage, and NF-kB signaling are associated with LGLL phenotype.
While current LGLL treatment protocols rely on immunosuppressive therapies, targeted therapies such as NF-κB inhibitors are under clinical investigations [1]. We demonstrated that DNMT inhibition can potently inhibit the STAT3-DNA methylation pathway in both CD8+ T cells of LGLL patients and KAI3 NK cells expressing activating STAT3 variants. Azacitidine, a therapeutic agent with DNA hypomethylating activity as the primary mechanism of action [53], inhibited ROS dependent NF-κB activation as well as STAT3 activation via restoration of SHP1, which is in line with a previous study using CD34 cells from AML patients [54] and anaplastic large cell lymphoma cells [55]. The negative correlation between SHP1 and p-STAT3 after azacitidine treatment in KAI3 NK cells is consistent with previous studies in other cell types [33, 56, 57]. SHP1 is primarily expressed in hematopoietic cells and is associated with DNA methylation and leukemogenesis [58]. As STAT3 can promote SHP1 gene silencing by forming complexes with DNMT1 and HDAC1 [49], the observed restoration of SHP1 expression by azacitidine might be explained by downregulated STAT3 activation. Conversely, siRNA targeting SHP1 was sufficient to abrogate the inhibitory effect of azacitidine on STAT3, suggesting that SHP1 is required for STAT3 inhibition by azacitidine treatment. The effect of azacitidine treatment on upregulating SHP-1 was confirmed in CD8+ T cells collected from a LGLL patient harboring STAT3Y640F mutation, suggesting that azacitidine may be used to target STAT3 mutated cells in LGLL patients. The treatment of LGL cells with demethylating drugs has also been shown to lead to upregulation of SOCS3 providing additional mechanism for observed STAT3 downregulation [13].
In conclusion, our results indicate that proinflammatory cytokine stimulation and STAT3 GOF mutations lead to epigenetic reprogramming, increased global DNA methylation, increased ROS production, and enhanced proliferation that may drive LGLL disease pathogenesis. The activity of this axis could be partially rescued by DNMT inhibition, providing novel targeted treatment options for LGLL.
References
Lamy T, Moignet A, Loughran TP Jr. LGL leukemia: from pathogenesis to treatment. Blood. 2017;129:1082–94.
Alekshun TJ, Tao J, Sokol L. Aggressive T-cell large granular lymphocyte leukemia: a case report and review of the literature. Am J Hematol. 2007;82:481–5.
Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl J Med. 2012;366:1905–13.
Andersson E, Kuusanmaki H, Bortoluzzi S, Lagstrom S, Parsons A, Rajala H, et al. Activating somatic mutations outside the SH2-domain of STAT3 in LGL leukemia. Leukemia. 2016;30:1204–8.
Barila G, Teramo A, Calabretto G, Vicenzetto C, Gasparini VR, Pavan L, et al. Stat3 mutations impact on overall survival in large granular lymphocyte leukemia: a single-center experience of 205 patients. Leukemia. 2020;34:1116–24.
Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng NgK, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120:3048–57.
Gasparini VR, Binatti A, Coppe A, Teramo A, Vicenzetto C, Calabretto G, et al. A high definition picture of somatic mutations in chronic lymphoproliferative disorder of natural killer cells. Blood Cancer J. 2020;10:42.
Rajala HL, Porkka K, Maciejewski JP, Loughran TP Jr., Mustjoki S. Uncovering the pathogenesis of large granular lymphocytic leukemia-novel STAT3 and STAT5b mutations. Ann Med. 2014;46:114–22.
Rajala HL, Olson T, Clemente MJ, Lagstrom S, Ellonen P, Lundan T, et al. The analysis of clonal diversity and therapy responses using STAT3 mutations as a molecular marker in large granular lymphocytic leukemia. Haematologica. 2015;100:91–9.
Savola P, Bruck O, Olson T, Kelkka T, Kauppi MJ, Kovanen PE, et al. Somatic STAT3 mutations in Felty syndrome: an implication for a common pathogenesis with large granular lymphocyte leukemia. Haematologica. 2018;103:304–12.
O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–70.
Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–62.
Teramo A, Gattazzo C, Passeri F, Lico A, Tasca G, Cabrelle A, et al. Intrinsic and extrinsic mechanisms contribute to maintain the JAK/STAT pathway aberrantly activated in T-type large granular lymphocyte leukemia. Blood. 2013;121:3843–54. S1
Moser B, Willimann K. Chemokines: role in inflammation and immune surveillance. Ann Rheum Dis. 2004;63:ii84–ii9. Suppl 2
Kang S, Tanaka T, Narazaki M, Kishimoto T. Targeting interleukin-6 signaling in clinic. Immunity 2019;50:1007–23.
Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185.
Mishra A, Liu S, Sams GH, Curphey DP, Santhanam R, Rush LJ, et al. Aberrant overexpression of IL-15 initiates large granular lymphocyte leukemia through chromosomal instability and DNA hypermethylation. Cancer Cell. 2012;22:645–55.
Fehniger TA, Suzuki K, Ponnappan A, VanDeusen JB, Cooper MA, Florea SM, et al. Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. J Exp Med. 2001;193:219–31.
Zambello R, Facco M, Trentin L, Sancetta R, Tassinari C, Perin A, et al. Interleukin-15 triggers the proliferation and cytotoxicity of granular lymphocytes in patients with lymphoproliferative disease of granular lymphocytes. Blood. 1997;89:201–11.
Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun. 2015;6:6025.
Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192.
Adnan AS, Dufva O, Ianevski A, Ghimire B, Koski J, Maliniemi P, et al. RUNX1 mutations in blast-phase chronic myeloid leukemia associate with distinct phenotypes, transcriptional profiles, and drug responses. Leukemia. 2021;35:1087–99.
Kim D, Park G, Huuhtanen J, Lundgren S, Khajuria RK, Hurtado AM, et al. Somatic mTOR mutation in clonally expanded T lymphocytes associated with chronic graft versus host disease. Nat Commun. 2020;11:2246.
Teramo A, Barila G, Calabretto G, Ercolin C, Lamy T, Moignet A, et al. STAT3 mutation impacts biological and clinical features of T-LGL leukemia. Oncotarget. 2017;8:61876–89.
Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9.
Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66.
Mishra A, La Perle K, Kwiatkowski S, Sullivan LA, Sams GH, Johns J, et al. Mechanism, consequences, and therapeutic targeting of abnormal IL15 signaling in cutaneous T-cell lymphoma. Cancer Discov. 2016;6:986–1005.
Mariotti B, Calabretto G, Rossato M, Teramo A, Castellucci M, Barila G, et al. Identification of a miR-146b-Fas ligand axis in the development of neutropenia in T large granular lymphocyte leukemia. Haematologica. 2020;105:1351–60.
Kreuz S, Fischle W. Oxidative stress signaling to chromatin in health and disease. Epigenomics. 2016;8:843–62.
Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc Natl Acad Sci USA. 2008;105:9302–6.
Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–15.
Brumell JH, Burkhardt AL, Bolen JB, Grinstein S. Endogenous reactive oxygen intermediates activate tyrosine kinases in human neutrophils. J Biol Chem. 1996;271:1455–61.
Huang TT, Su JC, Liu CY, Shiau CW, Chen KF. Alteration of SHP-1/p-STAT3 Signaling: A Potential Target for Anticancer Therapy. Int J Mol Sci. 2017;18.
Al-Jamal HA, Mat Jusoh SA, Hassan R, Johan MF. Enhancing SHP-1 expression with 5-azacytidine may inhibit STAT3 activation and confer sensitivity in lestaurtinib (CEP-701)-resistant FLT3-ITD positive acute myeloid leukemia. BMC Cancer. 2015;15:869.
Andersson EI, Brück O, Braun T, Mannisto S, Saikko L, Lagström S, et al. STAT3 mutation is associated with STAT3 activation in CD30+ ALK− ALCL. Cancers. 2020;12:702.
Greenplate A, Wang K, Tripathi RM, Palma N, Ali SM, Stephens PJ, et al. Genomic profiling of T-cell neoplasms reveals frequent JAK1 and JAK3 mutations with clonal evasion from targeted therapies. JCO Precis Oncol. 2018;2:PO.17.00019.
Dufva O, Kankainen M, Kelkka T, Sekiguchi N, Awad SA, Eldfors S, et al. Aggressive natural killer-cell leukemia mutational landscape and drug profiling highlight JAK-STAT signaling as therapeutic target. Nat Commun. 2018;9:1567.
Xiong J, Cui BW, Wang N, Dai YT, Zhang H, Wang CF, et al. Genomic and transcriptomic characterization of natural killer T cell lymphoma. Cancer Cell. 2020;37:403–19 e6.
Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Allen HL, De Franco E, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014;46:812–4.
Haapaniemi EM, Kaustio M, Rajala HL, van Adrichem AJ, Kainulainen L, Glumoff V, et al. Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood 2015;125:639–48.
Shvidel L, Duksin C, Tzimanis A, Shtalrid M, Klepfish A, Sigler E, et al. Cytokine release by activated T-cells in large granular lymphocytic leukemia associated with autoimmune disorders. Hematol J. 2002;3:32–7.
Kothapalli R, Nyland SB, Kusmartseva I, Bailey RD, McKeown TM, Loughran TP Jr. Constitutive production of proinflammatory cytokines RANTES, MIP-1beta and IL-18 characterizes LGL leukemia. Int J Oncol. 2005;26:529–35.
Chen J, Petrus M, Bamford R, Shih JH, Morris JC, Janik JE, et al. Increased serum soluble IL-15Ralpha levels in T-cell large granular lymphocyte leukemia. Blood 2012;119:137–43.
Wang TT, Yang J, Zhang Y, Zhang M, Dubois S, Conlon KC, et al. IL-2 and IL-15 blockade by BNZ-1, an inhibitor of selective gamma-chain cytokines, decreases leukemic T-cell viability. Leukemia. 2019;33:1243–55.
Wingelhofer B, Neubauer HA, Valent P, Han X, Constantinescu SN, Gunning PT, et al. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia. 2018;32:1713–26.
Dasgupta M, Dermawan JK, Willard B, Stark GR. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA. 2015;112:3985–90.
Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–52.
Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108:1058–64.
Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci USA. 2005;102:6948–53.
Battisti V, Maders LD, Bagatini MD, Santos KF, Spanevello RM, Maldonado PA, et al. Measurement of oxidative stress and antioxidant status in acute lymphoblastic leukemia patients. Clin Biochem. 2008;41:511–8.
Sallmyr A, Fan J, Rassool FV. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett. 2008;270:1–9.
Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49:1603–16.
Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13.
Fabre C, Grosjean J, Tailler M, Boehrer S, Ades L, Perfettini JL, et al. A novel effect of DNA methyltransferase and histone deacetylase inhibitors: NFkappaB inhibition in malignant myeloblasts. Cell Cycle. 2008;7:2139–45.
Han Y, Amin HM, Frantz C, Franko B, Lee J, Lin Q, et al. Restoration of shp1 expression by 5-AZA-2’-deoxycytidine is associated with downregulation of JAK3/STAT3 signaling in ALK-positive anaplastic large cell lymphoma. Leukemia. 2006;20:1602–9.
Fan LC, Teng HW, Shiau CW, Tai WT, Hung MH, Yang SH, et al. Pharmacological targeting SHP-1-STAT3 signaling is a promising therapeutic approach for the treatment of colorectal cancer. Neoplasia. 2015;17:687–96.
Rhee YH, Jeong SJ, Lee HJ, Lee HJ, Koh W, Jung JH, et al. Inhibition of STAT3 signaling and induction of SHP1 mediate antiangiogenic and antitumor activities of ergosterol peroxide in U266 multiple myeloma cells. BMC Cancer. 2012;12:28.
Oka T, Ouchida M, Koyama M, Ogama Y, Takada S, Nakatani Y, et al. Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res. 2002;62:6390–4.
Acknowledgements
The research was funded by the European Research Council (Project: M-IMM 647355), Academy of Finland Heal-Art consortium (314442) and special COVID funding (335527), ERA PerMed (JAKSTAT-TARGET consortium), Sigrid Juselius Foundation, Signe and Ane Gyllenberg Foundation, Helsinki Institute for Life Science, and Cancer Foundation Finland. R.M. was supported by the Austrian Science Fund (FWF) (SFB-F04707,SFB-F06105, and under the frame of ERA ERA-NET [I 4157-B]). Amplicon sequencing was performed at the Institute for Molecular Medicine Finland FIMM Technology Centre, which is supported by Biocenter Finland. We acknowledge IT Center for Science Ltd for computational resources. Finnish Red Cross Blood Service is acknowledged for providing healthy control samples.
Data sharing and declaration
RNA sequencing data are available from the corresponding author upon request. All other data are available in the Supplementary files.
Funding
Open access funding provided by University of Helsinki including Helsinki University Central Hospital.
Author information
Authors and Affiliations
Contributions
DK, MM, GP, and SM designed the study and experiments. JH, BG, and MK performed sequencing analysis. HR provided the clinical data and patient samples. RM provided important idea and critical comments on this study. WC provided study materials and input to study design. DK, MM, and SM wrote the manuscript with the input of other authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
SM has received honoraria and research funding from Novartis, Pfizer and Bristol-Myers Squibb (not related to this study). MM has received honoraria from Celgene and Sanofi. The remaining authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, D., Park, G., Huuhtanen, J. et al. STAT3 activation in large granular lymphocyte leukemia is associated with cytokine signaling and DNA hypermethylation. Leukemia 35, 3430–3443 (2021). https://doi.org/10.1038/s41375-021-01296-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01296-0
This article is cited by
-
Unlocking the secrets: the power of methylation-based cfDNA detection of tissue damage in organ systems
Clinical Epigenetics (2023)
-
Comprehensive pan-cancer analysis of STAT3 as a prognostic and immunological biomarker
Scientific Reports (2023)
-
Acetylcholine promotes chronic stress-induced lung adenocarcinoma progression via α5-nAChR/FHIT pathway
Cellular and Molecular Life Sciences (2023)
-
All that glitters is not LGL Leukemia
Leukemia (2022)
-
The lncRNA PRINS-miRNA-mRNA Axis Gene Expression Profile as a Circulating Biomarker Panel in Psoriasis
Molecular Diagnosis & Therapy (2022)