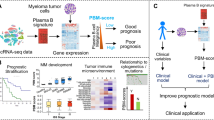
Abstract
Accurate survival prediction of persons with plasma cell myeloma (PCM) is challenging. We interrogated clinical and laboratory co-variates and RNA matrices of 1040 subjects with PCM from public datasets in the Gene Expression Omnibus database in training (N = 1) and validation (N = 2) datasets. Genes regulating plasma cell metabolism correlated with survival were identified and seven used to build a metabolic risk score using Lasso Cox regression analyses. The score had robust predictive performance with 5-year survival area under the curve (AUCs): 0.71 (95% confidence interval, 0.65, 0.76), 0.88 (0.67, 1.00) and 0.64 (0.57, 0.70). Subjects in the high‐risk training cohort (score > median) had worse 5-year survival compared with those in the low‐risk cohort (62% [55, 68%] vs. 85% [80, 90%]; p < 0.001). This was also so for the validation cohorts. A nomogram combining metabolic risk score with Revised International Staging System (R-ISS) score increased survival prediction from an AUC = 0.63 [0.58, 0.69] to an AUC = 0.73 [0.66, 0.78]; p = 0.015. Modelling predictions were confirmed in in vitro tests with PCM cell lines. Our metabolic risk score increases survival prediction accuracy in PCM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Gerecke C, Fuhrmann S, Strifler S, Schmidt-Hieber M, Einsele H, Knop S. The diagnosis and treatment of multiple myeloma. Dtsch Arztebl Int. 2016;113:470–6.
Hu Y, Chen W, Wang J. Progress in the identification of gene mutations involved in multiple myeloma. Onco Targets Ther. 2019;12:4075–80.
Flynt E, Bisht K, Sridharan V, Ortiz M, Towfic F, Thakurta A. Prognosis, biology, and targeting of TP53 dysregulation in multiple myeloma. Cells. 2020;9:287.
Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Blade J, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23:3412–20.
Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. Revised International Staging System for multiple myeloma: a report from International Myeloma Working Group. J Clin Oncol. 2015;33:2863–9.
Cho H, Yoon DH, Lee JB, Kim SY, Moon JH, Do YR, et al. Comprehensive evaluation of the revised international staging system in multiple myeloma patients treated with novel agents as a primary therapy. Am J Hematol. 2017;92:1280–6.
Xia B, Wang X, Yang R, Mengzhen L, Yang K, Ren L, et al. Epstein-Barr virus infection is associated with clinical characteristics and poor prognosis of multiple myeloma. Biosci Rep. 2019;39:BSR20190284.
Gonsalves WI, Jevremovic D, Nandakumar B, Dispenzieri A, Buadi FK, Dingli D, et al. Enhancing the R-ISS classification of newly diagnosed multiple myeloma by quantifying circulating clonal plasma cells. Am J Hematol. 2020;95:310–5.
Al Saleh AS, Sidiqi MH, Dispenzieri A, Kapoor P, Muchtar E, Buadi FK, et al. Hematopoietic score predicts outcomes in newly diagnosed multiple myeloma patients. Am J Hematol. 2020;95:4–9.
Jung SH, Kwon SY, Min JJ, Bom HS, Ahn SY, Jung SY, et al. F-FDG PET/CT is useful for determining survival outcomes of patients with multiple myeloma classified as stage II and III with the Revised International Staging System. Eur J Nucl Med Mol Imaging. 2019;46:107–15.
Bolli N, Genuardi E, Ziccheddu B, Martello M, Oliva S, Terragna C. Next-generation sequencing for clinical management of multiple myeloma: ready for prime time? Front Oncol. 2020;10:189.
EI Arfani C, De Veirman K, Maes K, De Bruyne E, Menu E. Metabolic features of multiple myeloma. Int J Mol Sci. 2018;19:1200.
Janker L, Mayer RL, Bileck A, Kreutz D, Mader JC, Utpatel K, et al. Metabolic, anti-apoptotic and immune evasion strategies of primary human myeloma cells indicate adaptations to hypoxia. Mol Cell Proteom. 2019;18:936–53.
Maiso P, Huynh D, Moschetta M, Sacco A, Aljawai Y, Mishima Y, et al. Metabolic signature identifies novel targets for drug resistance in multiple myeloma. Cancer Res. 2015;75:2071–82.
Hsu HC, Tong S, Zhou Y, Elmes MW, Yan S, Kaczocha M, et al. The antinociceptive agent SBFI-26 binds to anandamide transporters FABP5 and FABP7 at two different sites. Biochemistry. 2017;56:3454–62.
Higgins MJ, Graves PR, Graves LM. Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3. J Biol Chem. 2007;282:29493–503.
Helliwell SB, Karkare S, Bergdoll M, Rahier A, Leighton-Davis JR, Fioretto C, et al. FR171456 is a specific inhibitor of mammalian NSDHL and yeast Erg26p. Nat Commun. 2015;6:8613.
Liu XP, Yin XH, Meng XY, Yan XH, Wang F, He L. Development and validation of a 9-gene prognostic signature in patients with multiple myeloma. Front Oncol. 2018;8:615.
Berger WT, Ralph BP, Kaczocha M, Sun J, Balius TE, Rizzo RC, et al. Targeting fatty acid binding protein (FABP) anandamide transporters—a novel strategy for development of anti-inflammatory and anti-nociceptive drugs. PLoS One. 2012;7:e50968.
Kim DG, Cho S, Lee KY, Cheon SH, Yoon HJ, Lee JY, et al. Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity. Cell Mol Life Sci. 2020;78:207–25.
Li X, Yan X, Wang F, Yang Q, Luo X, Kong J, et al. Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. J Cell Mol Med. 2019;23:2920–32.
Rawat A, Gopal G, Selvaluxmy G, Rajkumar T. Inhibition of ubiquitin conjugating enzyme UBE2C reduces proliferation and sensitizes breast cancer cells to radiation, doxorubicin, tamoxifen and letrozole. Cell Oncol (Dordr). 2013;36:459–67.
Shafat MS, Oellerich T, Mohr S, Robinson SD, Edwards DR, Marlein CR, et al. Leukemic blasts program bone marrow adipocytes to generate a protumoral microenvironment. Blood 2017;129:1320–32.
Martin E, Minet N, Boschat AC, Sanquer S, Sobrino S, Lenoir C, et al. Impaired lymphocyte function and differentiation in CTPS1-deficient patients result from a hypomorphic homozygous mutation. JCI Insight. 2020;5:e133880.
De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Meyer C, Marschalek R, et al. FLNA, a new partner gene fused to MLL in a patient with acute myelomonoblastic leukaemia. Br J Haematol. 2009;146:693–5.
Palmer DC, Guittard GC, Franco Z, Crompton JG, Eil RL, Patel SJ, et al. Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance. J Exp Med. 2015;212:2095–113.
Acknowledgements
YL is supported, in part, by Sun Yat-sen University Start-up Funding, grant no. 201603, the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S096) and the National Natural Science Foundation of China (Grant No. 81873428). H-xL is supported by the National Natural Science Foundation of China (Grant No. 81773103) and the Natural Science Foundation of Guangdong Province (2017A030313617). JL is supported by Sun Yat-sen University Medical Clinical Trial ‘5010 Plan’ (2017005). RPG acknowledges support from the National Institute of Health Research (NIHR) Biomedical Research Centre funding scheme.
Author information
Authors and Affiliations
Contributions
H-yH and YW: preparing the typescript; YL, RPG, JL, H-yH and YW: reviewing and editing; W-dW and J-yL: software; X-lW and L-lS: methodology; Q-yZ and LL: conceptualisation; YL, H-xL and JL: design research, project administration and funding acquisition.
Corresponding authors
Ethics declarations
Conflict of interest
RPG is a paid Consultant to BeiGene Ltd., CStone Pharmaceuticals and Kite Pharmma; a Consultant to Fusion Pharma LLC, LaJolla NanoMedical Inc. and Mingsight Parmaceuticals Inc.; an Advisory Board member for Antegene Biotech LLC and StemRad Ltd; Medical Director at FFF Enterprises Inc; Partner in AZACA Inc; and on the Board of Directors of the Russian Foundation for Cancer Research Support. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Huang, Hy., Wang, Y., Wang, Wd. et al. A prognostic survival model based on metabolism-related gene expression in plasma cell myeloma. Leukemia 35, 3212–3222 (2021). https://doi.org/10.1038/s41375-021-01206-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01206-4
This article is cited by
-
Bone marrow stromal cells dictate lanosterol biosynthesis and ferroptosis of multiple myeloma
Oncogene (2024)
-
CTPS1 is a novel therapeutic target in multiple myeloma which synergizes with inhibition of CHEK1, ATR or WEE1
Leukemia (2024)
-
A novel NHEJ gene signature based model for risk stratification and prognosis prediction in hepatocellular carcinoma
Cancer Cell International (2023)
-
A novel prognostic model based on pyroptosis-related genes for multiple myeloma
BMC Medical Genomics (2023)
-
Integrated assessment of the clinical and biological value of ferroptosis-related genes in multiple myeloma
Cancer Cell International (2022)