Abstract
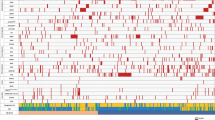
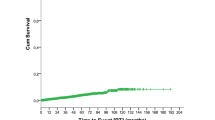
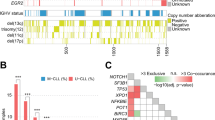
Large granular lymphocyte leukemia (LGLL) is a rare and chronic lymphoproliferative disorder characterized by the clonal expansion of LGLs. LGLL patients can be asymptomatic or develop cytopenia, mostly neutropenia. Somatic STAT3 and STAT5b mutations have been recently reported in approximately 40% of patients. The aim of this study is to analyze clinical and biological features of a large cohort of LGLL patients to identify prognostic markers affecting patients’ outcome. In 205 LGLL patients, neutropenia (ANC < 1500/mm3) was the main feature (38%), with severe neutropenia (ANC < 500/mm3) being present in 20.5% of patients. STAT3 mutations were detected in 28.3% patients and were associated with ANC < 500/mm3 (p < 0.0001), Hb < 90 g/L (p = 0.0079) and treatment requirement (p < 0.0001) while STAT5b mutations were found in 15/152 asymptomatic patients. By age-adjusted univariate analysis, ANC < 500/mm3 (p = 0.013), Hb < 90 g/L (p < 0.0001), treatment requirement (p = 0.001) and STAT3 mutated status (p = 0.011) were associated to reduced overall survival (OS). By multivariate analysis, STAT3 mutated status (p = 0.0089) and Hb < 90 g/L (p = 0.0011) were independently associated to reduced OS. In conclusion, we identified clinical and biological features associated to reduced OS in LGLL and we demonstrated the adverse impact of STAT3 mutations in patients’ survival, suggesting that this biological feature should be regarded as a potential target of therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zambello R, Semenzato G. Large granular lymphocyte disorders: new etiopathogenetic clues as a rationale for innovative therapeutic approaches. Haematologica. 2009;94:1341–5.
Steinway SN, LeBlanc F, Loughran TP Jr. The pathogenesis and treatment of large granular lymphocyte leukemia. Blood Rev. 2014;28:87–94.
Lamy T, Moignet A, Loughran TP Jr. LGL leukemia: from pathogenesis to treatment. Blood. 2017;129:1082–94.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon, France: IARC Press; 2017, 585pp.
Teramo A, Barila G, Calabretto G, Ercolin C, Lamy T, Moignet A, et al. STAT3 mutation impacts biological and clinical features of T-LGL leukemia. Oncotarget. 2017;8:61876–89.
Bareau B, Rey J, Hamidou M, Donadieu J, Morcet J, Reman O, et al. Analysis of a French cohort of patients with large granular lymphocyte leukemia: a report on 229 cases. Haematologica. 2010;95:1534–41.
Sanikommu SR, Clemente MJ, Chomczynski P, Afable MG 2nd, Jerez A, Thota S, et al. Clinical features and treatment outcomes in large granular lymphocytic leukemia (LGLL). Leuk Lymphoma. 2018;59:416–22.
Bockorny B, Dasanu CA. Autoimmune manifestations in large granular lymphocyte leukemia. Clin Lymphoma Myeloma Leuk. 2012;12:400–5.
Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–13.
Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng Ng K, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120:3048–57.
Rajala HL, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121:4541–50.
Andersson EI, Tanahashi T, Sekiguchi N, Gasparini VR, Bortoluzzi S, Kawakami T, et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood. 2016;128:2465–8.
Pandolfi F, Loughran TP Jr, Starkebaum G, Chisesi T, Barbui T, Chan WC, et al. Clinical course and prognosis of the lymphoproliferative disease of granular lymphocytes. A multicenter study. Cancer. 1990;65:341–8.
Lamy T, Loughran TP Jr. How I treat LGL leukemia. Blood. 2011;117:2764–74.
Shi M, He R, Feldman AL, Viswanatha DS, Jevremovic D, Chen D, et al. STAT3 mutation and its clinical and histopathologic correlation in T-cell large granular lymphocytic leukemia. Hum Pathol. 2018;73:74–81.
Bilori B, Thota S, Clemente MJ, Patel B, Jerez A, Afable Ii M, et al. Tofacitinib as a novel salvage therapy for refractory T-cell large granular lymphocytic leukemia. Leukemia. 2015;29:2427–9.
Kontro M, Kuusanmaki H, Eldfors S, Burmeister T, Andersson EI, Bruserud O, et al. Novel activating STAT5B mutations as putative drivers of T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:1738–42.
Nicolae A, Xi L, Pittaluga S, Abdullaev Z, Pack SD, Chen J, et al. Frequent STAT5B mutations in gammadelta hepatosplenic T-cell lymphomas. Leukemia. 2014;28:2244–8.
McKinney M, Moffitt AB, Gaulard P, Travert M, De Leval L, Nicolae A, et al. The genetic basis of hepatosplenic T-cell lymphoma. Cancer Discov. 2017;7:369–79.
Kiel MJ, Velusamy T, Rolland D, Sahasrabuddhe AA, Chung F, Bailey NG, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124:1460–72.
Lopez C, Bergmann AK, Paul U, Murga Penas EM, Nagel I, Betts MJ, et al. Genes encoding members of the JAK-STAT pathway or epigenetic regulators are recurrently mutated in T-cell prolymphocytic leukaemia. Br J Haematol. 2016;173:265–73.
Acknowledgements
The authors would like to thank Associazione Italiana per la Ricerca sul Cancro (AIRC, IG 2017-20216).
Author information
Authors and Affiliations
Contributions
GB designed the research, analyzed data and wrote the manuscript. AT, GC, CV and VRG performed mutations analysis. LP, ML and SV provided patient’s samples and patient’s data. MF contributed to analyze data. ACF performed statistical analysis. GS provided funding, participated in the analysis of data and critically reviewed and edited the manuscript. RZ designed the study, analyzed data, wrote the manuscript and supervised the study.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Barilà, G., Teramo, A., Calabretto, G. et al. Stat3 mutations impact on overall survival in large granular lymphocyte leukemia: a single-center experience of 205 patients. Leukemia 34, 1116–1124 (2020). https://doi.org/10.1038/s41375-019-0644-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0644-0
This article is cited by
-
The constitutive activation of STAT3 gene and its mutations are at the crossroad between LGL leukemia and autoimmune disorders
Blood Cancer Journal (2024)
-
Identification of novel STAT5B mutations and characterization of TCRβ signatures in CD4+ T-cell large granular lymphocyte leukemia
Blood Cancer Journal (2022)
-
Single-cell characterization of leukemic and non-leukemic immune repertoires in CD8+ T-cell large granular lymphocytic leukemia
Nature Communications (2022)
-
All that glitters is not LGL Leukemia
Leukemia (2022)
-
Defining TCRγδ lymphoproliferative disorders by combined immunophenotypic and molecular evaluation
Nature Communications (2022)