Abstract
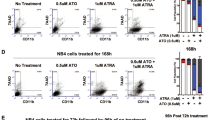
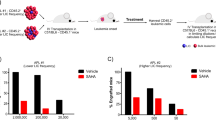
To date, only one subtype of acute myeloid leukemia (AML), acute promyelocytic leukemia (APL) can be effectively treated by differentiation therapy utilizing all-trans retinoic acid (ATRA). Non-APL AMLs are resistant to ATRA. Here we demonstrate that the acetyltransferase GCN5 contributes to ATRA resistance in non-APL AML via aberrant acetylation of histone 3 lysine 9 (H3K9ac) residues maintaining the expression of stemness and leukemia associated genes. We show that inhibition of GCN5 unlocks an ATRA-driven therapeutic response. This response is potentiated by coinhibition of the lysine demethylase LSD1, leading to differentiation in most non-APL AML. Induction of differentiation was not correlated to a specific AML subtype, cytogenetic, or mutational status. Our study shows a previously uncharacterized role of GCN5 in maintaining the immature state of leukemic blasts and identifies GCN5 as a therapeutic target in AML. The high efficacy of the combined epigenetic treatment with GCN5 and LSD1 inhibitors may enable the use of ATRA for differentiation therapy of non-APL AML. Furthermore, it supports a strategy of combined targeting of epigenetic factors to improve treatment, a concept potentially applicable for a broad range of malignancies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data discussed in this publication were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE124423.
Change history
16 January 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Longo DL, Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–52.
Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369:111–21.
de The H. Differentiation therapy revisited. Nat Rev Cancer. 2018;18:117–27.
Schenk T, Stengel S, Zelent A. Unlocking the potential of retinoic acid in anticancer therapy. Br J Cancer. 2014;111:2039–45.
Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteom. 2008;7:2019–27.
Glasow A, Barrett A, Petrie K, Gupta R, Boix-Chornet M, Zhou DC, et al. DNA methylation-independent loss of RARA gene expression in acute myeloid leukemia. Blood. 2008;111:2374–7.
Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–11.
Smitheman KN, Severson TM, Rajapurkar SR, McCabe MT, Karpinich N, Foley J, et al. Lysine specific demethylase 1 inactivation enhances differentiation and promotes cytotoxic response when combined with all-trans retinoic acid in acute myeloid leukemia across subtypes. Haematologica. 2018;104:1156–67.
Cusan M, Cai SF, Mohammad HP, Krivtsov A, Chramiec A, Loizou E, et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPalpha-dependent enhancers in AML. Blood. 2018;131:1730–42.
Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–87.
Mueller-Tidow C. Phase I/II Trial of ATRA and TCP in Patients With Relapsed or Refractory AML and no Intensive Treatment is Possible. https://ClinicalTrials.gov/show/NCT02261779. 2014.
Ross D. IMG-7289, With and Without ATRA, in Patients With Advanced Myeloid Malignancies. https://ClinicalTrials.gov/show/NCT02842827. 2016.
Lübbert M. Study of Sensitization of Non-M3 AML Blasts to ATRA by Epigenetic Treatment With Tranylcypromine (TCP). https://ClinicalTrials.gov/show/NCT02717884. 2016.
Zheng F. An Open-Label, Dose-Escalation/Dose-Expansion Safety Study of INCB059872 in Subjects With Advanced Malignancies. https://ClinicalTrials.gov/show/NCT02712905. 2016.
Watts J. Phase 1 Study of TCP-ATRA for Adult Patients With AML and MDS. https://ClinicalTrials.gov/show/NCT02273102. 2014.
Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–900.
Kuo Y-M, Andrews AJ. Quantitating the Specificity and Selectivity of Gcn5-Mediated Acetylation of Histone H3. PLoS ONE. 2013;8:e54896.
Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016;17:1193–205.
Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9.
Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–37.
Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sonderby CK, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917–24.
Biel M, Kretsovali A, Karatzali E, Papamatheakis J, Giannis A. Design, Synthesis, and Biological Evaluation of a Small‐Molecule Inhibitor of the Histone Acetyltransferase Gcn5. Angew Chem Int Ed. 2004;43:3974–6.
Dalton WT Jr., Ahearn MJ, McCredie KB, Freireich EJ, Stass SA, et al. HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3. Blood. 1988;71:242–7.
Picot T, Aanei CM, Fayard A, Flandrin-Gresta P, Tondeur S, Gouttenoire M, et al. Expression of embryonic stem cell markers in acute myeloid leukemia. Tumour Biol. 2017;39:1010428317716629.
Xu H, Huang S, Zhu X, Zhang W, Zhang X. FOXK1 promotes glioblastoma proliferation and metastasis through activation of Snail transcription. Exp Ther Med. 2018;15:3108–16.
Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708.
Rice KL, Licht JD. HOX deregulation in acute myeloid leukemia. J Clin Investig. 2007;117:865–8.
Santanach A, Blanco E, Jiang H, Molloy KR, Sanso M, LaCava J, et al. The Polycomb group protein CBX6 is an essential regulator of embryonic stem cell identity. Nat Commun. 2017;8:1235.
Salvatori B, Iosue I, Mangiavacchi A, Loddo G, Padula F, Chiaretti S, et al. The microRNA-26a target E2F7 sustains cell proliferation and inhibits monocytic differentiation of acute myeloid leukemia cells. Cell Death Dis. 2012;3:e413.
Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell. 2016;165:303–16.
Ramirez RN, El-Ali NC, Mager MA, Wyman D, Conesa A, Mortazavi A. Dynamic gene regulatory networks of human myeloid differentiation. Cell Syst. 2017;4:416–29. e413.
Vegi NM, Klappacher J, Oswald F, Mulaw MA, Mandoli A, Thiel VN, et al. MEIS2 is an oncogenic partner in AML1-ETO-positive AML. Cell Rep. 2016;16:498–507.
Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci USA. 2005;102:11728–33.
Gal H, Amariglio N, Trakhtenbrot L, Jacob-Hirsh J, Margalit O, Avigdor A, et al. Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia. 2006;20:2147–54.
Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–93.
Lin LI, Chen CY, Lin DT, Tsay W, Tang JL, Yeh YC, et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11:1372–9.
Martelli MP, Pettirossi V, Thiede C, Bonifacio E, Mezzasoma F, Cecchini D, et al. CD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised mice. Blood. 2010;116:3907–22.
Boutzen H, Saland E, Larrue C, de Toni F, Gales L, Castelli FA, et al. Isocitrate dehydrogenase 1 mutations prime the all-trans retinoic acid myeloid differentiation pathway in acute myeloid leukemia. J Exp Med. 2016;213:483–97.
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9.
Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–6.
Bararia D, Kwok H, Welner RS, Numata A, Sárosi MB, Yang H, et al. Acetylation of C/EBPα inhibits its granulopoietic function. Nat Commun. 2016;7:10968.
Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer activation by pharmacologic displacement of lsd1 from gfi1 induces differentiation in acute myeloid leukemia. Cell Rep. 2018;22:3641–59.
Cain C. AML takes LSD1. Science-Business eXchange. 2012;5:352.
Fu X, Zhang P, Yu B. Advances toward LSD1 inhibitors for cancer therapy. Future Med Chem. 2017;9:1227–42.
Acknowledgements
We are grateful to Maren Godmann, Berit Jungnickel, and Christian Kosan for their advice during the project. We thank Ivonne Goerlich and Cornelia Luge for technical assistance in next-generation sequencing, Carl Crodel, Kai Sporkmann, and Tobias Rachow for their help collecting patients’ material and Carl Götze for his advice in stochastic analyses. MK and TS were supported by the German Research Council (SCHE1909/2–1). AB was supported by funding from the Foundation “Else Kröner-Fresenius-Stiftung”. FHH was supported by grants of the Thuringian state program ProExzellenz (RegenerAging—FSU-I-03/14) of the Thuringian Ministry for Research and in part by the German Research Council (DFG, HE6233/6–1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kahl, M., Brioli, A., Bens, M. et al. The acetyltransferase GCN5 maintains ATRA-resistance in non-APL AML. Leukemia 33, 2628–2639 (2019). https://doi.org/10.1038/s41375-019-0581-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0581-y