Abstract
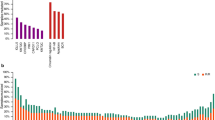
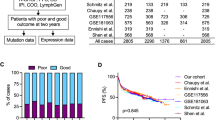
Diffuse large B-cell lymphoma (DLBCL) is a biologically and clinically heterogeneous disease whose personalized clinical management requires robust molecular stratification. Here, we show that somatic hypermutation (SHM) patterns constitute a marker for DLBCL molecular classification. The activity of SHM mutational processes delineated the cell of origin (COO) in DLBCL. Expression of the herein identified 36 SHM target genes stratified DLBCL into four novel SHM subtypes. In a meta-analysis of patients with DLBCL treated with immunochemotherapy, the SHM subtypes were significantly associated with overall survival (1642 patients) and progression-free survival (795 patients). Multivariate analysis of survival indicated that the prognostic impact of the SHM subtypes is independent from the COO classification and the International Prognostic Index. Furthermore, the SHM subtypes had a distinct clinical outcome within each of the COO subtypes, and strikingly, even within unclassified DLBCL. The genetic landscape of the four SHM subtypes indicated unique associations with driver alterations and oncogenic signaling in DLBCL, which suggests a possibility for therapeutic exploitation. These findings provide a biologically driven classification system in DLBCL with potential clinical applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Coiffier B, Lepage E, Brie’re J, Herbrecht R, Tilly H, Bouabdallah R, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. New Engl J Med. 2002;346:235–42.
Pfreundschuh M, Trümper L, Österborg A, Pettengell R, Trneny M, Imrie K, et al. CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in youngpatients with good-prognosis diffuse large-B-cell lymphoma: a randomised controlled trialby the MabThera International Trial (MInT) Group. Lancet Oncol. 2006;7:379–91.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000 feb;403:503–11.
Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-Cell lymphoma. New Engl J Med. 2002;346:1937–47.
Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. New Engl J Med. 2008;359:2313–23.
Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–94. e15
Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–65.
Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109:3879–84.
Dubois S, Viailly PJ, Mareschal S, Bohers E, Bertrand P, Ruminy P, et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA Study. Clin Cancer Res. 2016;22:2919–28.
Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. New Engl J Med. 2018;378:1396–407.
Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018 may;24:679–90.
Arthur SE, Jiang A, Grande BM, Alcaide M, Cojocaru R, Rushton CK, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun. 2018;9:4001.
Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti R, Küppers R, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–6.
Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, et al. Twolevels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–5.
Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4(5):452–6.
Khodabakhshi AH, Morin RD, Fejes AP, Mungall AJ, Mungall KL, Bolger-Munro M, et al. Recurrent targets of aberrant somatic hypermutation in lymphoma. Oncotarget. 2012;3:1308.
Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, et al. AID is required for germinal center–derived lymphomagenesis. Nat Genet. 2008;40:108.
Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–61.
Dybkær K, Bøgsted M, Falgreen S, Bødker JS, Kjeldsen MK, Schmitz A, et al. Diffuse large B-Cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J Clin Oncol. 2015;33:1379–88.
Ennishi D, Jiang A, Boyle M, Collinge B, Grande BM, Ben-Neriah S, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol. 2019;37:190–201.
Sha C, Barrans S, Cucco F, Bentley MA, Care MA, Cummin T, et al. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J Clin Oncol. 2019;37:202–12.
Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181.
Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103–13.
Icay K, Chen P, Cervera A, Rantanen V, Lehtonen R, Hautaniemi S. SePIA: RNA and small RNA sequence processing, integration, and analysis. BioData Min. 2016;9:20.
Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat methods. 2013;10:71–73.
Cervera A, Rantanen V, Ovaska K, Laakso M, Nuñez-Fontarnau J, Alkodsi A, et al. Anduril 2: Upgraded large-scale data integration framework. Bioinformatics. 2019;btz133.
Ovaska K, Laakso M, Haapa-Paananen S, Louhimo R, Chen P, Aittomäki V, et al. Large-scaledata integration framework provides a comprehensive view on glioblastoma multiforme. Genome Med. 2010;2:65.
Supek F, Lehner B. Clustered mutation signatures reveal that error-prone DNA repair targets mutations to active genes. Cell. 2017;170:534–47.
Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinforma. 2010;11:367.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3.
Kapp AV, Tibshirani R. Are clusters found in one dataset present in another dataset? Biostatistics. 2006;8:9–31.
Alexandrov LB, Nik-Zainal S, Wedge DC. Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G: C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172:3382–4.
Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–7.
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90.
Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J Immunol. 2001;166:5051–7.
Meriranta L, Pasanen A, Louhimo R, Cervera A, Alkodsi A, Autio M, et al. Deltex-1 mutations predict poor survival in diffuse large B-cell lymphoma. Haematologica. 2017;102:e195–e198.
Acknowledgements
This work was supported financially by the Academy of Finland (SH, SL), the Sigrid Jusélius Foundation (SH, SL), Finnish Cancer Foundations (SH, SL), Helsinki University Hospital (SL), the Biomedicum Helsinki Foundation (AA) and the University of Helsinki graduate program (AA). The results published here are in part based upon data generated by the Cancer Genome Characterization Initiative (Non-Hodgkin Lymphoma project), and data generated by the Genomic Variation in Diffuse Large B Cell Lymphoma study, which was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services. Both datasets have been accessed through the NIH database for Genotypes and Phenotypes (dbGAP) with accession numbers phs000532 and phs001444. Information about CGCI projects can be found at https://ocg.cancer.gov/programs/cgci. The authors thank Dr. Ryan D Morin and Dr. Sandeep Dave for providing gene expression data. The international DLBCL consortia whose data we have used in this study are gratefully acknowledged. The authors thank professors Ville Mustonen and Jussi Taipale for critical review of the article. Computing resources from CSC – IT Center for Science Ltd. and technical assistance from Anne Aarnio and Marika Tuukkanen are gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
AA conceptualized the study together with R Lehtonen, SL, and SH; AA designed the methodology, performed the analyses, made the figures and wrote the paper; AA, AC, KZ, and R Louhimo processed sequencing data; AP, SKL, LM, HH, and SL provided resources and materials for the in-house samples and participated in scientific discussions; SH, SL, and R Lehtonen supervised the study; SH and SL acquired funding; all authors read and edited the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alkodsi, A., Cervera, A., Zhang, K. et al. Distinct subtypes of diffuse large B-cell lymphoma defined by hypermutated genes. Leukemia 33, 2662–2672 (2019). https://doi.org/10.1038/s41375-019-0509-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0509-6
This article is cited by
-
Activation-induced cytidine deaminase causes recurrent splicing mutations in diffuse large B-cell lymphoma
Molecular Cancer (2024)
-
Clinical implications of circulating tumor DNA in predicting the outcome of diffuse large B cell lymphoma patients receiving first-line therapy
BMC Medicine (2022)
-
Circulating RNA biomarkers in diffuse large B-cell lymphoma: a systematic review
Experimental Hematology & Oncology (2021)