Abstract
We have previously shown that dividing patients with CLL into those with telomeres inside the fusogenic range (TL-IFR) and outside the fusogenic range (TL-OFR) is powerful prognostic tool. Here, we used a high-throughput version of the assay (HT-STELA) to establish whether telomere length could predict for outcome to fludarabine, cyclophosphamide, rituximab (FCR)-based treatment using samples collected from two concurrent phase II studies, ARCTIC and ADMIRE (n = 260). In univariate analysis, patients with TL-IFR had reduced progression-free survival (PFS) (P < 0.0001; HR = 2.17) and shorter overall survival (OS) (P = 0.0002; HR = 2.44). Bifurcation of the IGHV-mutated and unmutated subsets according to telomere length revealed that patients with TL-IFR in each subset had shorter PFS (HR = 4.35 and HR = 1.48, respectively) and shorter OS (HR = 3.81 and HR = 2.18, respectively). In addition, the OS of the TL-OFR and TL-IFR subsets were not significantly altered by IGHV mutation status (P = 0.61; HR = 1.24 and P = 0.41; HR = 1.47, respectively). In multivariate modeling, telomere length was the dominant co-variable for PFS (P = 0.0002; HR = 1.85) and OS (P = 0.05; HR = 1.61). Taken together, our data suggest that HT-STELA is a powerful predictor of outcome to FCR-based treatment and could be used to inform the design of future risk-adapted clinical trials.
Similar content being viewed by others
Introduction
The management of chronic lymphocytic leukemia (CLL) has undergone a dramatic change in the last decade due to the introduction of effective new therapeutic regimens. However, despite the demonstrable efficacy of B-cell receptor signaling antagonists like ibrutinib [1, 2], the gold standard treatment for fit patients with CLL is still fludarabine, cyclophosphamide, rituximab (FCR) combination chemoimmunotherapy [3,4,5,6,7]. However, longer follow-up studies have shown that not all patients benefit equally from FCR; IGHV-mutated patients have a significantly better overall survival compared to IGHV-unmutated patients [5, 8,9,10] and patients with 17p abnormalities show markedly inferior outcomes with this regimen [8,9,10].
We and others have previously shown that telomeres play a pivotal role in the pathogenic progression and outcome of CLL, with telomere length analysis providing independent prognostication in all stages of CLL [11,12,13,14]. Using high-resolution telomere length analysis (STELA), we were able to show that patients with very short, dysfunctional telomeres are prone to chromosome fusion events; these patients commonly demonstrate increased genomic complexity [11, 14]. Furthermore, many of the losses and gains of genetic material observed in these patients appear to be focused at chromosome-ends and are consistent with the types of genetic rearrangements that can occur because of telomere fusion, leading to the initiation of cycles of anaphase-bridging, breakage, and fusion [15]. Importantly, we have shown previously that telomere length is the strongest determinant of clinical outcome in CLL and predicts for survival following chemotherapy [16, 17].
Here, we investigated telomere length in the context of frontline treatment with fludarabine, cyclophosphamide, rituximab (FCR)-based regimens. We considered whether telomere length was predictive of clinical outcome, using high-throughput single telomere length analysis (HT-STELA), in 275 samples from two concurrent UK trials of FCR-based treatment, ARCTIC and ADMIRE. We then compared the predictive power of HT-STELA with the commonly used prognostic markers: β2 microglobulin (β2M), fluorescence in situ hybridization (FISH) cytogenetics, CD38 expression, ZAP70 expression and IGHV mutation status.
Materials and methods
All the clinical samples used were taken at study entry and were obtained from the Bloodwise funded UK CLL Trials Biobank, University of Liverpool. Of the 260 evaluable samples for telomere length, 128 were derived from the ARCTIC trial; 64 patients were randomized to receive standard dose fludarabine, cyclophosphamide and rituximab (FCR) and 64 received fludarabine, cyclophosphamide, mitoxantrone, and mini rituximab (FCM-mini R). One-hundred thirty-two samples were evaluated from the the ADMIRE study; 64 patients were randomized to receive standard dose FCR and 68 received fludarabine, cyclophosphamide, mitoxantrone and rituximab (FCMR). 17p mutation or deletion were exclusion criteria from both of these studies due to their association with poor outcome following FCR treatment [18, 19]. However, due to the lag time in genetic analysis, it was later established that 16 patients with a 17p deletion were enrolled in the trials. The median follow-up in the combined cohort was 4 years and there were 51 deaths at the censor point. The demographics of the cohort are summarized in Table 1. Due to the study inclusion criteria for ARCTIC and ADMIRE, disease burden was generally high with a mean absolute lymphocyte count of 87.6 × 106/mL (range 3.3–547.5). However, to avoid potential measurement error caused by the presence of non-malignant cell fractions, telomere length was assessed on DNA extracted from purified CD19+ B-cells using a B-cell isolation kit (Miltenyi Biotec) using an adaptation of chromosome-specific STELA to allow for high-throughput analysis (HT-STELA). Briefly, the previously published STELA protocol [20, 21] was adapted to use telomere-adjacent primers specific for the XpYp telomere (XpYpC: 5ʹ-CAGGGACCGGGACAAATAGAC-3ʹ) and the 7q telomere (7qK1: 5ʹ-GGGCACTGCCTCGCTTTGA-3ʹ), in triplicate 30 μL PCR reactions each containing 30 ng of genomic DNA. Thermal cycling conditions were: 23 cycles of 94 °C for 20 s, 65 °C for 30 s, and 68 °C for 5 min. Amplified fragments were resolved using capillary gel electrophoresis and mean telomere length determined using PROSize software (AATI, Ankeny, Iowa, USA). Patients were bifurcated using the previously determined mean XpYp telomere length threshold for telomere dysfunction [17], creating two patient groups: one with telomere lengths equal or less than the mean of the fusogenic range; inside the fusogenic range (TL-IFR) and the other with telomere lengths greater than the mean of the fusogenic range; outside the fusogenic range (TL-OFR). The numerical threshold that defined these two groups using XpYp telomere analysis was subsequently adjusted for the 7q telomere according to the y = mx + c regression line generated by plotting XpYp telomere length against 7q telomere length. 7q HT-STELA was used in preference to XpYp HT-STELA as a larger subset of CLL samples failed to amplify the XpYp telomere (24/275) when compared with the 7q telomere (15/275). For consistency, all of the subsequent analyses were carried out on the data generated using 7q HT-STELA (n = 260). However, it should be noted that there was a strong correlation between the telomere lengths at each chromosome end (P < 0.0001; r2 = 0.72). In univariate analyses, all the individual prognostic markers were considered as categorical variables using previously established thresholds. In multivariate modeling, the same parameters were evaluated as both categorical and continuous variables. The variables included in the model were age, cytogenetic groups (del 11q and del 17p), IGHV mutation status, CD38 expression, ZAP70 expression, β2M, absolute lymphocyte count, telomere length. Statistical analysis was carried out using Prism 6.0 (Graphpad Software Inc., La Jolla, CA, USA) and SAS version 9.3 software (SAS Institute, Cary, NC, USA). Univariate comparisons for progression-free survival (PFS) and overall survival (OS) were conducted with the logrank test and displayed as Kaplan-Meier curves. Multivariate analyses were performed using a Cox proportional hazard model with forward selection. In all cases P < 0.05 was considered significant.
Results
High-throughtput STELA allows for the reliable and rapid evaluation of telomere length in CLL
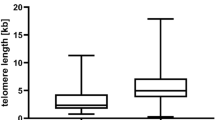
Our previously described single molecule STELA assay is both technically challenging and time consuming making it unsuitable for the evaluation of large numbers of samples [11]. To overcome these problems, we developed a modification of the STELA assay to facilitate the high-throughput evaluation of samples (HT-STELA). Here we present the first evidence that this technique is comparable to standard STELA and can be used to quickly and reliably predict for outcome following FCR-based therapy in samples derived from two UK CLL trials, ARCTIC and ADMIRE [6, 7]. To evaluate the utility of HT-STELA for the analysis of telomere length in CLL, we undertook a comparison of both STELA and HT-STELA on 260 patient samples, at two separate chromosome-ends using primers designed to specifically amplify the XpYp and the 7q telomeres. We showed strong concordance between the STELA and HT-STELA assays; Fig. 1a shows 7q STELA versus 7q HT-STELA (r2 = 0.92, P < 0.0001). In addition, Fig. 1b shows the correlation between the XpYp telomere length and the 7q telomere length using HT-STELA (r2 = 0.72, P < 0.0001). It is worthy of note that the variance between the two chromosome-ends was most apparent in samples with long telomeres and no sample changed category (TL-IFR, TL-OFR) as a consequence of measuring a different telomere. Consistent with previous reports [11, 14, 15, 21], short telomere length was significantly associated with CD38 positivity (P = 0.0002, Fig. 1c) and elevated β2M (P = 0.038, Fig. 1d), IGHV-unmutated cases (P < 0.0001, Fig. 1e) and high-risk cytogenetic lesions (P < 0.0001, Fig. 1f).
Telomere length measurements using HT-STELA and the relationship between telomere length and other prognostic markers. HT-STELA was developed to overcome some of the technical complexities of the original STELA assay and to adapt the assay to make it suitable for high-throughput analysis. a shows the strong correlation between original STELA and HT-STELA. b shows the strong correlation between telomere length measured using HT-STELA with probes specific for the XpYp and 7q telomeres. c–f 7q HT-STELA confirmed that poor prognostic subsets all had significantly shorter telomere length profiles than their respective good prognosis counter-parts
Telomere length predicts for progression-free and overall survival
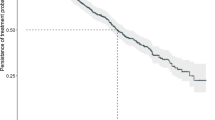
We first set out to establish whether telomere length could predict for outcome to FCR-based therapy in the entire cohort of samples, regardless of randomization. The cohort was split according to our previously defined threshold of telomere dysfunction; telomere lengths equal or less than the mean of the fusogenic range (TL-IFR) and telomere lengths greater than the mean of the fusogenic range (TL-OFR). Patients with TL-IFR showed significantly shorter PFS (P < 0.0001; Fig. 2a) and reduced OS (P = 0.0002; Fig. 2b). In the same cohort, IGHV mutation status was predictive of PFS (P = 0.0016; Fig. 2c) but not OS (P = 0.38; Fig. 2d). Bifurcation of the IGHV-mutated and IGHV-unmutated groups into TL-IFR and TL-OFR subsets showed that telomere length identified patients with different PFS (Fig. 2e) and different OS (Fig. 2f). Neither CD38 expression (< / > 20%) nor β2M (< / > 3.5 mg/L) expression were predictive of PFS or OS (Supplementary Fig. 1A–D). In addition, removal of the 17p deleted/mutated patients from the analysis (n = 16) did not significantly alter the predictive power of telomere length in terms of PFS and OS (Supplementary Fig. 2). Although the 7q HT-STELA analysis was selected for use in this study, XpYp HT-STELA showed similar predictive power for both PFS and OS (Supplementary Fig. 3).
Stratification of patients by telomere length predicts for PFS and OS following FCR-based treatment. Bifurcation of the patient cohort according to the previously defined telomere length threshold for telomere dysfunction was predictive of (a) PFS and (b) OS. Patients whose telomere length were ≤ the mean of the fusogenic range (TL-IFR) showed shorter PFS and OS than those patients with mean telomere length outside of the fusogenic range (TL-OFR). Comparison of the same cohort based on IGHV mutation status showed that IGHV-unmutated patients had c a shorter median PFS but no difference in d OS when compared with IGHV-mutated patients. IGHV-mutated and IGHV-unmutated groups could be divided according to telomere length, which identified distinct subsets in each group with e different PFS and f different OS
Telomere length identifies differential outcomes in IGHV-mutated and IGHV-unmutated groups
A number of studies, including the German CLL8 study, have shown that a proportion of IGHV-mutated patients, treated with FCR, can achieve long-term remissions [8,9,10]. Here, we show that patients with IGHV-mutated CLL with short telomeres (TL-IFR) were more likely to progress during the follow-up period (P < 0.0001, HR = 4.35; Fig. 3a) and more likely to succumb to their disease (P = 0.006, HR = 3.81; Fig. 3c) than patients with TL-OFR. In the IGHV-unmutated group, most patients showed clinical progression during the follow-up period; the median PFS of of the TL-IFR subset was 3.7 years versus 4.83 years in the TL-OFR subset (P = 0.08, HR = 1.48; Fig. 3b). The IGHV-unmutated TL-IFR subset showed a significantly worse OS when compared with the TL-OFR subset (P = 0.025, HR = 2.18; Fig. 3d). It is of interest that the IGHV-mutated subset with TL-IFR had a median OS almost one year less than the IGHV-unmutated subset with TL-IFR (4.49 years versus 5.47 years). Although the number of IGHV-mutated patients with TL-IFR was relatively small (n = 16), our data suggests that telomere length can identify a subset of “bad risk” IGHV-mutated patients who do not respond well to FCR. Furthermore, splitting the cohort into TL-IFR and TL-OFR subsets was a more powerful determinant of clinical outcome than IGHV mutation status. Indeed, the OS of patients with TL-OFR and TL-IFR were not significantly different regardless of IGHV mutation status (Fig. 3e, f respectively). It is worthy of note that of the 16 “bad risk” IGHV-mutated patients only 2/16 used the IGHV 3–21 gene. In terms of other known prognostic factors, 5/16 were 11q-, 2/16 were 17p-, 10/16 were CD38+ and 11/16 had β2M ≥3.5 mg/L.
The impact of telomere length in IGHV-mutated and IGHV-unmutated groups. Dividing the IGHV-mutated and IGHV-unmutated prognostic groups according to telomere length revealed a a significant difference in PFS in the IGHV-mutated group with short telomere patients having shorter PFS. b Short telomere length in the IGHV-unmutated group showed a trend towards reduced PFS but this was not significant. In terms of OS, the short telomere subsets in both c IGHV-mutated and d IGHV-unmutated groups showed significantly reduced survival. The OS of patients with e long telomeres (TL-OFR) and f short telomeres (TL-IFR) were not significantly different regardless of IGHV mutation status
Telomere length is predictive in both ARCTIC and ADMIRE regardless of randomization
We next analyzed the effect of telomere length in the samples derived from the two separate trials and between the randomization arms of the two studies. We showed that there was no difference in telomere length between patients randomized to receive FCR and FCMR in the ADMIRE study (P = 0.92; Fig. 4a) and those randomized to receive FCR and FCM-mini R in the ARCTIC study (P = 0.59; Fig. 4b). In both studies, telomere length analysis was able to identify subsets of patients with different PFS (Fig. 4c, d) and different OS (Fig. 4e, f) to the FCR-based treatments given.
Comparison of telomere length in patients treated in ADMIRE and ARCTIC. The mean telomere length were not significantly different between patients randomized in a ADMIRE and b ARCTIC clinical trials. Stratification accord to telomere length revealed differential PFS (c and d) and OS (e and f) in both clinical trials regardless of randomization
Telomere length is predictive in patients treated with standard dose FCR
Although we demonstrated that mean telomere length was not different between the patients randomized within both ADMIRE and ARCTIC, the published data on the two trials suggested a randomization effect [6, 7] so we performed a sub-analysis of the effect of telomere length in all patients treated with standard dose FCR (n = 136). Patients with short telomeres who received FCR showed a significantly shorter PFS (P = 0.0014, HR = 2.4; Fig. 5a) and shorter OS (P = 0.0037, HR = 2.91; Fig. 5b). In the same cohort, IGHV mutation status was predictive of PFS (P = 0.05, HR = 1.79; Fig. 5c) but not OS (P = 0.36, HR = 1.47; Fig. 5d). Once again, telomere length was able to identify a subset of both IGHV-mutated and IGHV-unmutated cases with a differential PFS and OS when treated with FCR (Fig. 5e, f respectively). Consistent with the analysis of the entire cohort, the IGHV-mutated cases with TL-IFR showed a shorter median PFS (2.53 years) and a shorter median OS (4.49 years) than the IGHV-unmutated cases with TL-IFR (3.61 years and 4.81 years respectively; Supplementary Fig. 4).
The impact of telomere length in FCR-treated patients derived from ADMIRE and ARCTIC. Analysis of the patient cohort treated with standard dose FCR demonstrated that telomere length was a strong predictor of a PFS and b OS. Comparison of the effect of IGHV mutation status in the same cohort showed it was c predictive of PFS but d was not predictive of OS. Bifurcation of the IGHV-mutated and IGHV-unmutated groups accord to telomere length again reveals subsets with distinct e PFS and f OS
Telomere length is the dominant co-variable in multivariate analysis
In univariate analysis we have shown that telomere length is a strong predictor of PFS and OS in FCR-based clinical trials. However, given the association between short telomeres and other high-risk prognostic markers, we next set out to determine whether telomere length would retain predictive significance in multivariate analysis using a Cox proportional hazard model with forward selection. Table 2A and 2B summarize the univariate and multivariate analysis for PFS and OS; telomere length was the dominant co-variable with HR = 1.85 (1.33–2.28) and HR = 1.61 (0.99–2.63) respectively. Indeed, the inclusion of telomere length in the model rendered all of the other markers insignificant even when the inclusion threshold was reduced to P = 0.1. The tables show the adjusted P-values for telomere length when all of the other parameters were forced into the model and the unadjusted P-values for each parameter without forcing. In keeping with the previously published data from the ARCTIC and ADMIRE studies [6, 7], age was not an independent prognostic factor in either univariate or multivariate analysis.
Discussion
It has been previously shown that a proportion of chronic lymphocytic leukemia (CLL) patients have critically short telomeres and this is associated with genomic instability and inferior clinical outcome [11, 14]. Indeed, telomere dysfunction was shown to be the most powerful predictor of survival in a cohort of 321 Patients with CLL and allowed the accurate stratification of Binet stage A patients into those with indolent disease and those with poor prognosis [17]. Furthermore, patients with long telomeres showed superior prognosis regardless of their IGHV mutation status CD38 expression, ZAP70 expression or cytogenetic risk group. In keeping with this finding, telomere dysfunction was the dominant variable in multivariate analysis. More recently we proved that telomere dysfunction predicted for response to the combination of fludarabine and cyclophosphamide in the UK CLL4 clinical trial [16]. Here, we describe for the first time the utility of a high-throughput adaptation of single telomere length analysis (HT-STELA), in 260 evaluable samples derived from the ARCTIC and ADMIRE clinical trials; both trials evaluated the chemoimmunotherapy combination of fludarabine, cyclophosphamide, rituximab (FCR) in previously untreated Patients with CLL.
We showed that HT-STELA could be reliably used to determine telomere length at two different chromosome-ends. Our data also confirmed our previous findings that telomere length measurement at any chromosome end is a bellwether of the telomere length of all chromosome-ends in the absecnce of genetic lesions that lead to the loss of telomeric material. Shorter telomere length was associated CD38 positivity, increased β2 microglobulin, IGHV-unmutated gene sequences and high-risk cytogenetics. Division of the cohort according to the previously determined mean telomere length threshold for telomere dysfunction [18] revealed that telomere length is a powerful predictor of both PFS and OS in patients treated with FCR-based therapies. In contrast, CD38 expression and β2 microglobulin expression were not predictive and IGHV mutation status was only predictive of PFS. It remains unclear why our findings are somewhat different to the German CLL8 study in terms of OS but the longer median follow-up in the CLL8 study (5.9 years versus 4 years) may be a contributing factor as there appears to be a steepening in the FCR-treated IGHV-unmutated survival curve after 5 years [8]. In any case, telomere length is able to identify good risk and bad risk patients in both the IGHV-mutated and IGHV-unmutated groups (Figs. 2e, f and 3). Furthermore, telomere length appears to identify a subset of IGHV-mutated cases with a particularly poor response to FCR-based treatment; these patients had a worse median PFS and median OS than their IGHV-unmutated counter-parts. Furthermore, the adverse impact of short telomeres could not be explained by IGHV gene usage (IGHV 3–21) [22] or high-risk cytogenetic lesions (11q-/17p-). Although this finding is potentially important, it would need to be confirmed in a larger study as the number of IGHV-mutated cases with short telomeres was small (n = 16) when compared with the IGHV-unmutated subset (n = 60). It is of particular note that telomere length appears to be a critical determinant of clinical outcome and superseded the prognostic impact of IGHV mutation status in this cohort of patients (Fig. 3e, f).
Since the samples analyzed in this study were drawn from two separate clinical trials, ADMIRE and ARCTIC, we next evaluated the influence of telomere length in each trial separately and within each of the respective trial arms. There was no significant difference in mean telomere in either arm of the trials but telomere length was able to identify good risk and bad risk groups regardless of randomization (Fig. 4). Subsequent analysis of only the FCR-treated samples drawn from both trials (n = 136) revealed a similar picture to the entire cohort analysis. Telomere length was predictive of PFS and OS and could bifurcate IGHV-mutated and IGHV-unmutated groups in terms of PFS and OS (Fig. 5 and Supplementary Fig. 2).
Finally, we undertook multivariate modeling to establish the predictive hierarchy of the prognostic factors measured in this study. Using a forward selection model, telomere length was shown to be the dominant co-variable for both PFS and OS (Table 2A and B, respectively). The strong correlation between short telomeres and other poor-risk prognostic tools, including IGHV mutation status, appeared to render these markers insignificant when telomere length was included. Given the relative speed and simplicity of HT-STELA, our data suggest that it can reliably identify patients who will benefit from FCR treatment and those who will not. It remains to be established whether HT-STELA can also be used to predict for response to non-chemotherapeutic agents like ibrutinib but it seems likely that adoption of this predictive biomarker into clinical trials design could enable rational, risk-adapted, trials with the aim of treating all patients with the optimal therapeutic regimen.
References
Byrd JC, Furman RR, Coutre SE, Burger JA, Blum KA, Coleman M, et al. Three-year follow-up of treatment-naïve and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125:2497–506.
Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42.
Tam CS, O’Brien S, Wierda W, Kantarjian H, Wen S, Do KA, et al. Long-term results of the fludarabine, cyclophosphamide, and rituximab regimen as initial therapy of chronic lymphocytic leukemia. Blood. 2008;112:975–80.
Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–74.
Eichhorst B, Fink AM, Bahlo J, Busch R, Kovacs G, Maurer C, et al. First-line chemoimmunotherapy with bendamustine and rituximab versus fludarabine, cyclophosphamide, and rituximab in patients with advanced chronic lymphocytic leukaemia (CLL10): an international, open-label, randomised, phase 3, non-inferiority trial. Lancet Oncol. 2016;17:928–42.
Munir T, Howard DR, McParland L, Pocock C, Rawstron AC, Hockaday A, et al. Results of the randomized phase IIB ADMIRE trial of FCR with or without mitoxantrone in previously untreated CLL. Leukemia. 2017;31:2085–93.
DR Howard, T Munir, L McParland, AC Rawstron, D Milligan, A Schuh, et al. Results of the randomized phase IIB ARCTIC trial of low dose Rituximab in previously untreated CLL. Leukemia. 2017. https://doi.org/10.1038/leu.2017.96. [Epub ahead of print]
Fischer K, Bahlo J, Fink AM, Goede V, Herling CD, Cramer P, et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127:208–15.
Rossi D, Terzi-di-Bergamo L, De Paoli L, Cerri M, Ghilardi G, Chiarenza A, et al. Molecular prediction of durable remission after first-line fludarabine-cyclophosphamide-rituximab in chronic lymphocytic leukemia. Blood. 2015;126:1921–4.
Thompson PA, Tam CS, O’Brien SM, Wierda WG, Stingo F, Plunkett W, et al. Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood. 2016;127:303–9.
Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–907.
Pepper C, Majid A, Lin TT, Hewamana S, Pratt G, Walewska R, et al. Defining the prognosis of early stage chronic lymphocytic leukaemia patients. Br J Haematol. 2012;156:499–507.
Rossi D, Lobetti Bodoni C, Genuardi E, Monitillo L, Drandi D, Cerri M, et al. Telomere length is an independent predictor of survival, treatment requirement and Richter’s syndrome transformation in chronic lymphocytic leukemia. Leukemia. 2009;23:1062–72.
Roos G, Kröber A, Grabowski P, Kienle D, Bühler A, Döhner H, et al. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood. 2008;111:2246–52.
Murnane JP. Telomere dysfunction and chromosome instability. Mutat Res. 2012;730:28–36.
Strefford JC, Kadalayil L, Forster J, Rose-Zerilli MJ, Parker A, Lin TT, et al. Telomere length predicts progression and overall survival in chronic lymphocytic leukemia: data from the UK LRF CLL4 trial. Leukemia. 2015;29:2411–4.
Lin TT, Norris K, Heppel NH, Pratt G, Allan JM, Allsup DJ, et al. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br J Haematol. 2014;167:214–23.
Kovacs G, Robrecht S, Fink AM, Bahlo J, Cramer P, von Tresckow J, et al. Minimal residual disease assessment improves prediction of outcome in patients with chronic lymphocytic leukemia (CLL) who achieve partial response: comprehensive analysis of two phase III studies of the German CLL Study Group. J Clin Oncol. 2016;34:3758–65.
Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Döhner K, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123:3247–54.
Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–508.
Britt-Compton B, Lin TT, Ahmed G, Weston V, Jones RE, Fegan C, et al. Extreme telomere erosion in ATM-mutated and 11q-deleted Patients with CLL is independent of disease stage. Leukemia. 2012;26:826–30.
Thorsélius M, Kröber A, Murray F, Thunberg U, Tobin G, Bühler A, et al. Strikingly homologous immunoglobulin gene rearrangements and poor outcome in VH3-21-using chronic lymphocytic leukemia patients independent of geographic origin and mutational status. Blood. 2006;107:2889–94.
Acknowledgements
This work was funded in part by a Bloodwise Specialist Program Grant (12049) and the Wales Cancer Research Center. The study was undertaken with the support of the UK CLL Clinical Trials Sub-Group. Patient material was obtained from the UK CLL Trials Biobank, University of Liverpool, which is funded by Bloodwise.
Author contributions
KN carried out experiments, analyzed data, and revised the manuscript, PH provided vital reagents and revised the manuscript, AR provided vital reagents and revised the manuscript. RH analyzed data, CP conceived the study, analyzed data, and wrote the manuscript, DMB conceived the study, analyzed data, and revised the manuscript, CDF conceived the study, analyzed data, and revised the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
KN, CP, DMB, and CDF are all co-inventors of patents relevant to the content of this manuscript and hold shares in a University spin-out company set up to provide telomere length testing. PH, AR, and RH declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Norris, K., Hillmen, P., Rawstron, A. et al. Telomere length predicts for outcome to FCR chemotherapy in CLL. Leukemia 33, 1953–1963 (2019). https://doi.org/10.1038/s41375-019-0389-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0389-9
This article is cited by
-
Experimental and Computational Approaches to Measure Telomere Length: Recent Advances and Future Directions
Current Hematologic Malignancy Reports (2023)
-
Combined analysis of IGHV mutations, telomere length and CD49d identifies long-term progression-free survivors in TP53 wild-type CLL treated with FCR-based therapies
Leukemia (2022)
-
High-throughput STELA provides a rapid test for the diagnosis of telomere biology disorders
Human Genetics (2021)