Abstract
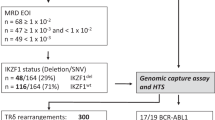
Deregulated expression of the type I cytokine receptor, CRLF2, is observed in 5–15% of precursor B-cell acute lymphoblastic leukaemia (B-ALL). We have previously reported the genomic landscape of patients with CRLF2 rearrangements (CRLF2-r) using both whole genome and exome sequencing, which identified a number of potential clonal and sub-clonal genomic alterations. In this study, we aimed to assess when the CRLF2-r; IGH-CRLF2 or P2RY8-CRLF2, arose during the evolution of both Down syndrome-ALL (DS-ALL) and non-DS-ALL. Using fluorescence in situ hybridisation, we were able to track up to four structural variants in single cells from 47 CRLF2-r B-ALL patients, which in association with our multiplex single-cell analysis of a further four patients, permitted simultaneous tracking of copy number alterations, structural and single nucleotide variants within individual cells. We observed CRLF2-r arising as both early and late events in DS and non-DS-ALL patients. Parallel evolution of discrete clones was observed in the development of CRLF2-r B-ALL, either involving the CRLF2-r or one of the other tracked abnormalities. In-depth single-cell analysis identified both linear and branching evolution with early clones harbouring a multitude of abnormalities, including the CRLF2-r in DS-ALL patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
References
Moorman AV. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. 2012;26:123–35.
Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–503.
Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–33.
Maia AT, van der Velden VHJ, Harrison CJ, Szczepanski T, Williams MD, Griffiths MJ, et al. Prenatal origin of hyperdiploid acute lymphoblastic leukemia in identical twins. Leukemia. 2003;17:2202–6.
Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci USA. 2002;99:8242–7.
Wiemels JL, Ford AM, Van Wering ER, Postma A, Greaves M. Protracted and variable latency of acute lymphoblastic leukemia AfterTEL-AML1 gene fusion in utero. blood. 1999;94:1057–62.
Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–34.
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LAA, Miller CB, et al. Deletion of IKZF1 and Prognosis in Acute Lymphoblastic Leukemia. New Engl J Med. 2009;360:470–80.
Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. New Engl J Med. 2014;371:1005–15.
Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114:2688–98.
Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–6.
Chapiro E, Russell L, Lainey E, Kaltenbach S, Ragu C, Della-Valle V, et al. Activating mutation in the TSLPR gene in B-cell precursor lymphoblastic leukemia. Leukemia. 2010;24:642–5.
Moorman AV, Schwab C, Ensor HM, Russell LJ, Morrison H, Jones L, et al. IGH@ Translocations, CRLF2 Deregulation, and Microdeletions in Adolescents and Adults With Acute Lymphoblastic Leukemia. J Clin Oncol. 2012;30:3100–8.
Schwab CJ, Jones LR, Morrison H, Ryan SL, Yigittop H, Schouten JP, et al. Evaluation of multiplex ligation-dependent probe amplification as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:1104–13.
Buitenkamp TD, Pieters R, Gallimore NE, van der Veer A, Meijerink JPP, Beverloo HB, et al. Outcome in children with Down’s syndrome and acute lymphoblastic leukemia: role of IKZF1 deletions and CRLF2 aberrations. Leukemia. 2012;26:2204–11.
Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–66.
Morak M, Attarbaschi A, Fischer S, Nassimbeni C, Grausenburger R, Bastelberger S, et al. Small sizes and indolent evolutionary dynamics challenge the potential role of P2RY8-CRLF2–harboring clones as main relapse-driving force in childhood ALL. Blood. 2012;120:5134–42.
Schwartzman O, Savino AM, Gombert M, Palmi C, Cario G, Schrappe M, et al. Suppressors and activators of JAK-STAT signaling at diagnosis and relapse of acute lymphoblastic leukemia in Down syndrome. Proc Natl Acad Sci USA. 2017;114:E4030–E9.
Wolman SR. Cytogenetic heterogeneity: its role in tumor evolution. Cancer Genet Cytogenet. 1986;19:129–40.
Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–61.
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604.
Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–25.
Potter NE, Ermini L, Papaemmanuil E, Cazzaniga G, Vijayaraghavan G, Titley I, et al. Single-cell mutational profiling and clonal phylogeny in cancer. Genome Res. 2013;23:2115–25.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4.
Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New Engl J Med. 2012;366:883–92.
Russell LJ, Jones L, Enshaei A, Tonin S, Ryan SL, Eswaran J, et al. Characterisation of the genomic landscape of CRLF2-rearranged acute lymphoblastic leukemia. Genes Chromosomes Canc. 2017;56:363–72.
le Viseur C, Hotfilder M, Bomken S, Wilson K, Rottgers S, Schrauder A, et al. In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell. 2008;14:47–58.
Jeffries SJ, Jones L, Harrison CJ, Russell LJ. IGH@ translocations co-exist with other primary rearrangements in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2014;99:1334–42.
Furness CL, Mansur MB, Weston VJ, Ermini L, van Delft FW, Jenkinson S, et al. The subclonal complexity of STIL-TAL1+T-cell acute lymphoblastic leukaemia. Leukemia. 2018;32:1984–1993.
De Bie J, Demeyer S, Alberti-Servera L, Geerdens E, Segers H, Broux M, et al. Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia. 2018;321:358–1369.
Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33.
Dussiau C, Fontenay M. Mechanisms underlying the heterogeneity of myelodysplastic syndromes. Exp Hematol. 2018;58:17–26.
Kim T, Tyndel MS, Kim HJ, Ahn JS, Choi SH, Park HJ, et al. The clonal origins of leukemic progression of myelodysplasia. Leukemia. 2017;31:1928–35.
Kent DG, Ortmann CA, Green AR. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372:1865–6.
Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–80.
van Delft FW, Horsley S, Colman S, Anderson K, Bateman C, Kempski H, et al. Clonal origins of relapse in ETV6-RUNX1 acute lymphoblastic leukemia. Blood. 2011;117:6247–54.
Ford AM, Mansur MB, Furness CL, van Delft FW, Okamura J, Suzuki T, et al. Protracted dormancy of pre-leukemic stem cells. Leukemia. 2015;29:2202–7.
Melchor L, Brioli A, Wardell CP, Murison A, Potter NE, Kaiser MF, et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28:1705–15.
Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–33.
Davies NJ, Kwok M, Gould C, Oldreive CE, Mao J, Parry H, et al. Dynamic changes in clonal cytogenetic architecture during progression of chronic lymphocytic leukemia in patients and patient-derived murine xenografts. Oncotarget. 2017;8:44749–60.
Belderbos ME, Koster T, Ausema B, Jacobs S, Sowdagar S, Zwart E, et al. Clonal selection and asymmetric distribution of human leukemia in murine xenografts revealed by cellular barcoding. Blood. 2017;129:3210–20.
Schmitz M, Breithaupt P, Scheidegger N, Cario G, Bonapace L, Meissner B, et al. Xenografts of highly resistant leukemia recapitulate the clonal composition of the leukemogenic compartment. Blood. 2011;118:1854–64.
Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47:257–62.
Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell . 2011;144:27–40.
Hart JR, Zhang Y, Liao L, Ueno L, Du L, Jonkers M, et al. The butterfly effect in cancer: a single base mutation can remodel the cell. Proc Natl Acad Sci USA. 2015;112:1131–6.
Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–8.
Cabelof DC, Patel HV, Chen Q, van Remmen H, Matherly LH, Ge Y, et al. Mutational spectrum at GATA1 provides insights into mutagenesis and leukemogenesis in Down syndrome. Blood. 2009;114:2753–63.
Hertzberg L, Vendramini E, Ganmore I, Cazzaniga G, Schmitz M, Chalker J, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood 2010;115:1006-17.
Acknowledgements
The authors would like to thank The Kay Kendall Leukaemia Fund, Leuka, Children with Cancer UK and Bloodwise (formerly Leukaemia and Lymphoma Research) for financial support. Lisa J Russell has a John Goldman Fellowship from Leuka. We also thank member laboratories of the United Kingdom Cancer Cytogenetic Group (UKCCG) for providing cytogenetic data and material. We are grateful to all the members of the NCRI Haematological Oncology Adult ALL Subgroup and the NCRI Childhood Cancer and Leukaemia Group (CCLG) Leukaemia Subgroup. Primary childhood leukaemia samples used in this study were provided by the Bloodwise Childhood Leukaemia Cell Bank working with the laboratory teams in the Bristol Genetics Laboratory, Southmead Hospital, Bristol: Molecular Biology Laboratory, Royal Hospital for Sick Children, Glasgow: Molecular Haematology Laboratory, Royal London Hospital, London: Molecular Genetics Service and Sheffield Children’s Hospital, Sheffield. We also thank the Central England Haemato-Oncology Research Biobank for providing patient samples. Finally, we thank all the clinicians who entered patients into the trial and the patients and families who agreed to take part.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Potter, N., Jones, L., Blair, H. et al. Single-cell analysis identifies CRLF2 rearrangements as both early and late events in Down syndrome and non-Down syndrome acute lymphoblastic leukaemia. Leukemia 33, 893–904 (2019). https://doi.org/10.1038/s41375-018-0297-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0297-4
This article is cited by
-
BCR::ABL1-like acute lymphoblastic leukaemia: a single institution experience on identification of potentially therapeutic targetable cases
Molecular Cytogenetics (2023)
-
Germinal Center-Related G Protein-Coupled Receptors in Antibody-Mediated Autoimmune Skin Diseases: from Basic Research to Clinical Trials
Clinical Reviews in Allergy & Immunology (2022)
-
RAS-protein activation but not mutation status is an outcome predictor and unifying therapeutic target for high-risk acute lymphoblastic leukemia
Oncogene (2021)
-
Emerging molecular subtypes and therapeutic targets in B-cell precursor acute lymphoblastic leukemia
Frontiers of Medicine (2021)
-
Alantolactone inhibits cell autophagy and promotes apoptosis via AP2M1 in acute lymphoblastic leukemia
Cancer Cell International (2020)